library(tidyverse)
theme_set(theme_bw())
cols <- c(
"gender", "name", "weight", "height", "method"
)
transport <- "data/transport.csv" |>
read_csv(
comment = "#",
col_names = cols,
col_types = "-ccnnc"
) |>
mutate(
gender = case_when(
gender == "F" ~ "female",
gender == "M" ~ "male",
TRUE ~ str_to_lower(gender)
),
gender = as.factor(gender),
method = factor(
method,
levels = c("car", "bike")
),
BMI = weight / (0.01 * height) ^ 2
)Putting It All Together: Advanced Plotting
Introduction to R For Biologists and Bioinformatics
Dr Stevie Pederson
Black Ochre Data Labs
Telethon Kids Institute
September 19, 2023
Using the
Tidyverse
Together
Using the Tidyverse
We’ve already learned a huge amount:
- Importing Data using
readr - Making plots with
ggplot2 - Working with vectors and logical tests
- Using
dplyrwith spreadsheet-like data - Joining functions together with
|>
- Next we’ll:
- Use everything to make more detailed figures
- Perform a few analyses
- Utilise some features of
tidyr
Section Setup
- Clear your
REnvironment - Start a new
Rscript:AdvancedPlotting.R - Load the
tidyverse - Define a default
ggplot2theme - Import Transport
Section Outline
- Using the pipe to create plots
- Customising plots in detail
- Reshaping data using
tidyr
Visualising Data
Visualising Our Data
- What might we like to show?
- Relationship between weight & height?
- Association with transportation method
- Distributions of BMI?
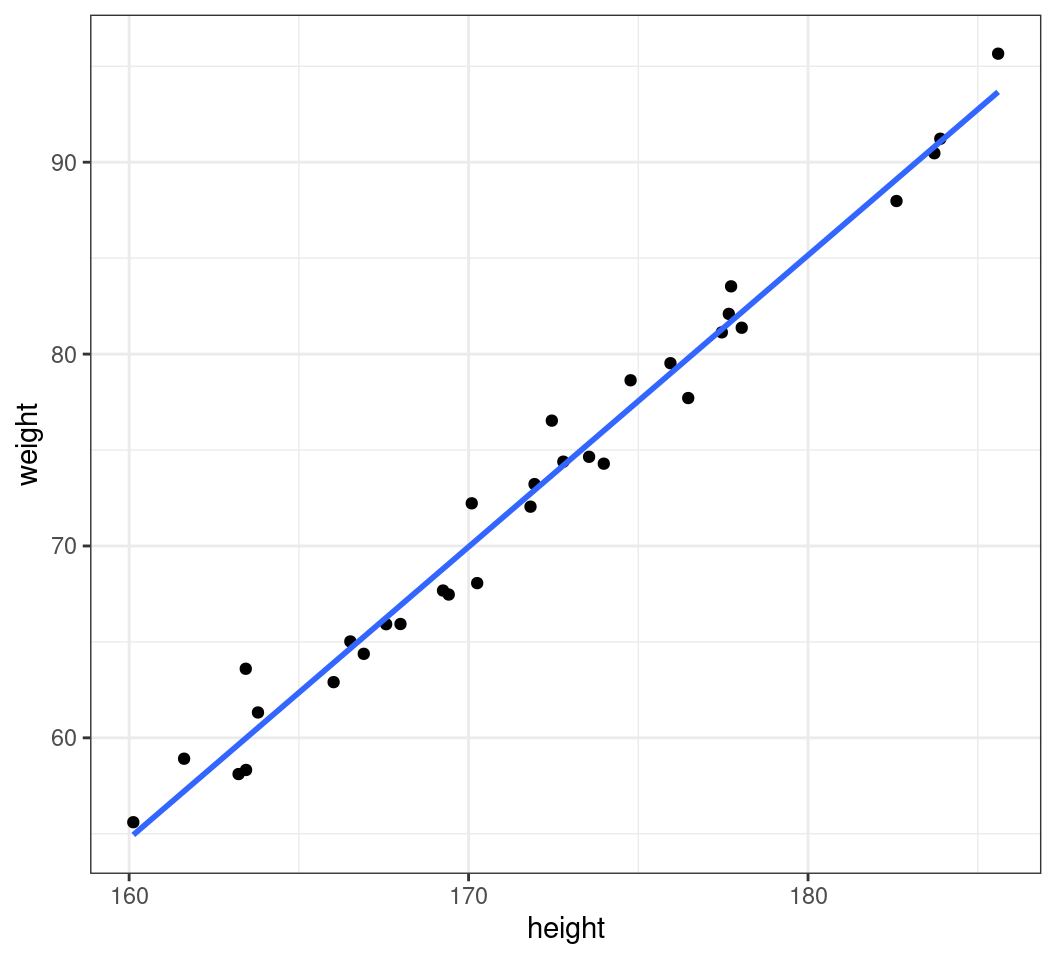
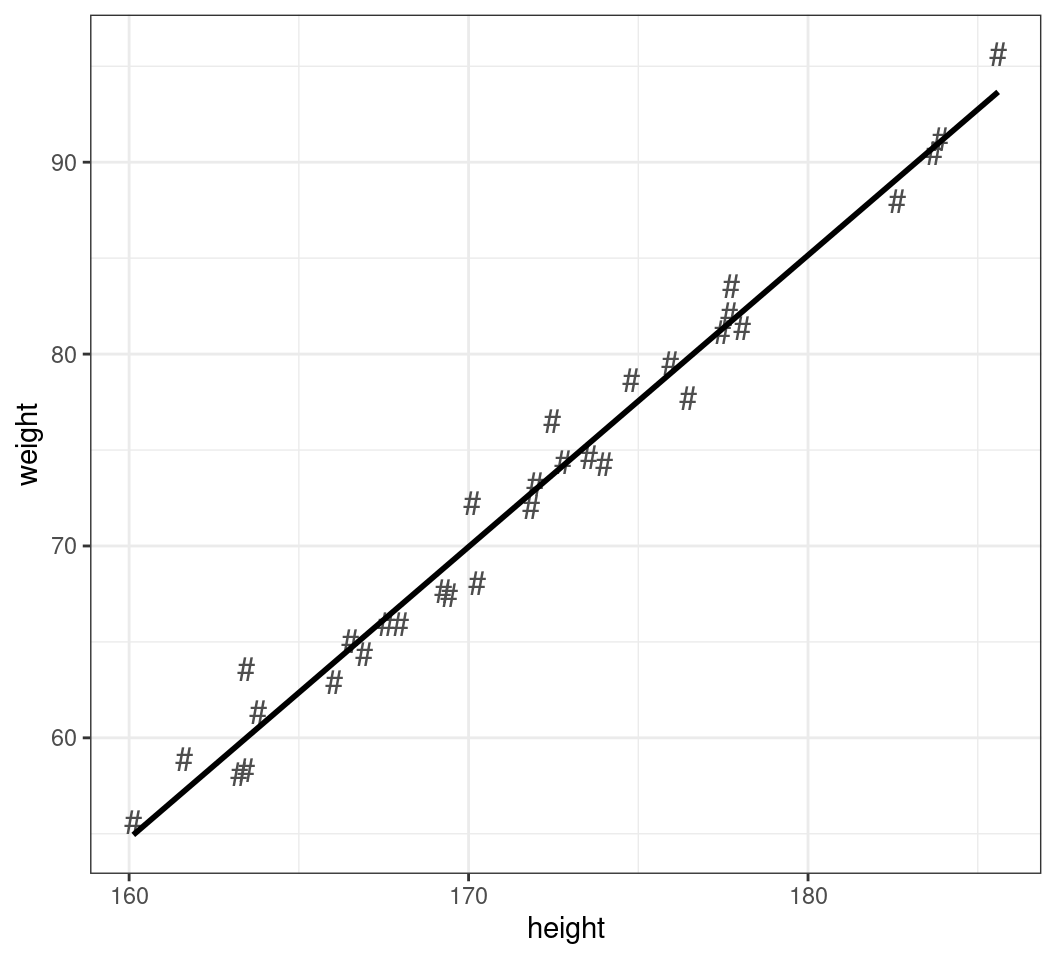
A Simple X-Y Plot
- When plotting, we can simply pipe the data into
ggplot
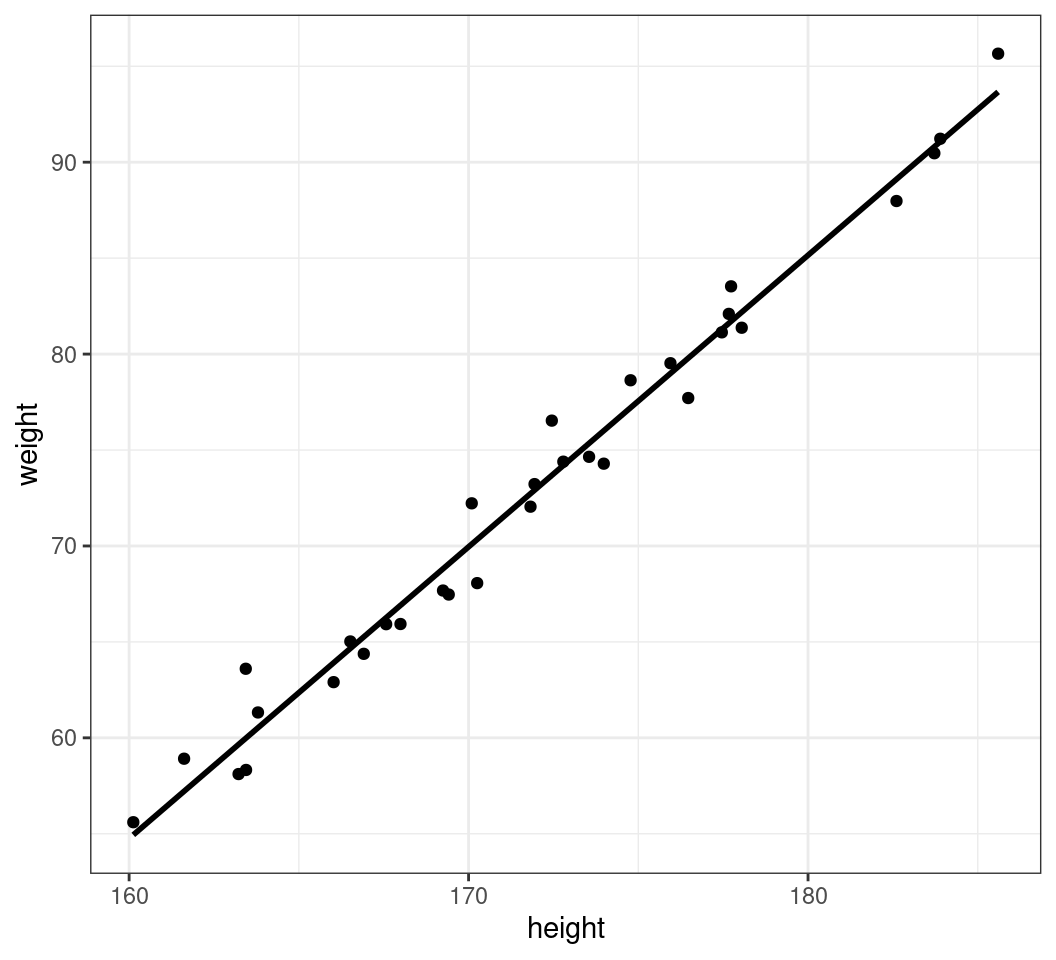
Adding Lines of Best Fit
- Sometimes a regression line can be informative
geom_smooth()guesses the best linemethod = 'loess'isn’t that great here
Adding Regresion Lines
- We can choose a linear regression line:
method = "lm"- We fit linear regression in
Rusing the functionlm() - Hide the standard error of the regression line:
se = FALSE
- We fit linear regression in
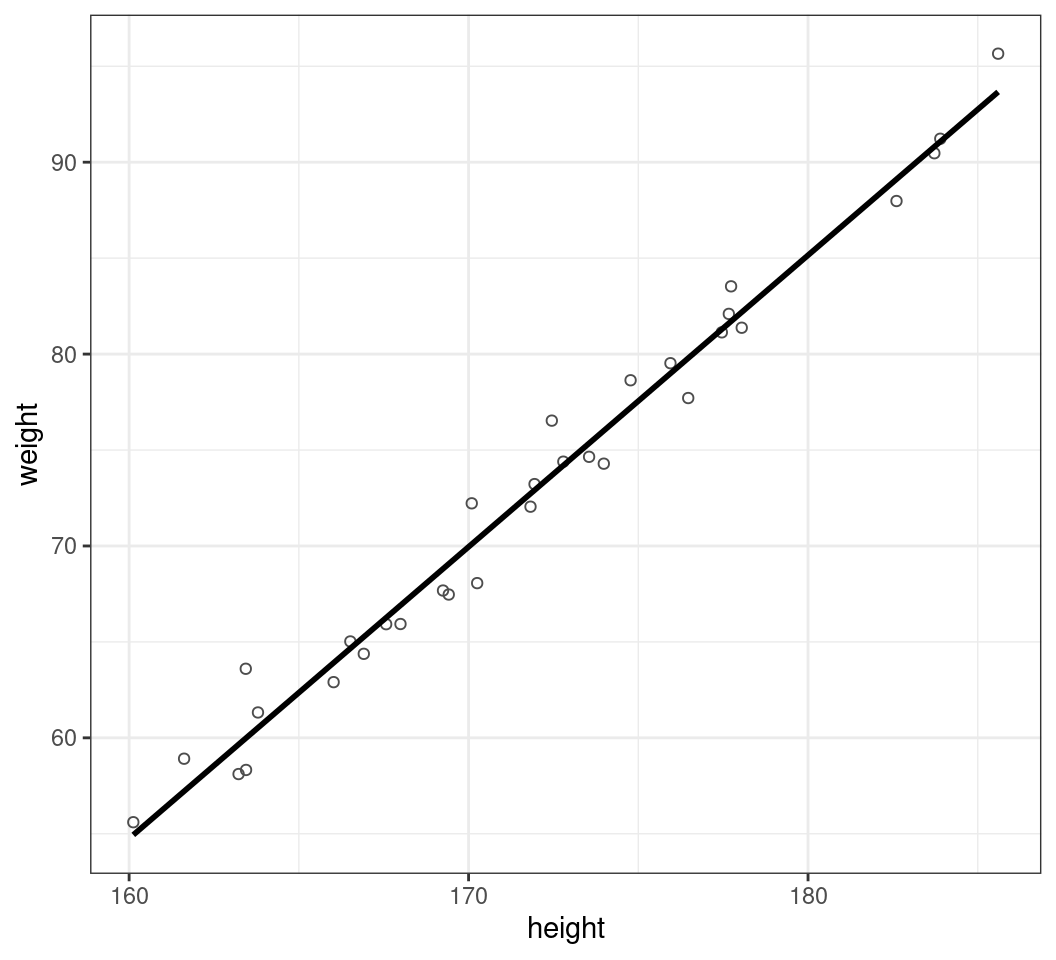
Customising Parameters
- We can change the colour of the line:
colour = "black"- Anything set inside
aes()should be a column in your data - Anything set outside of
aes()should be a fixed-value - Can also set
linetype,linewidth,alphaetc
- Anything set inside
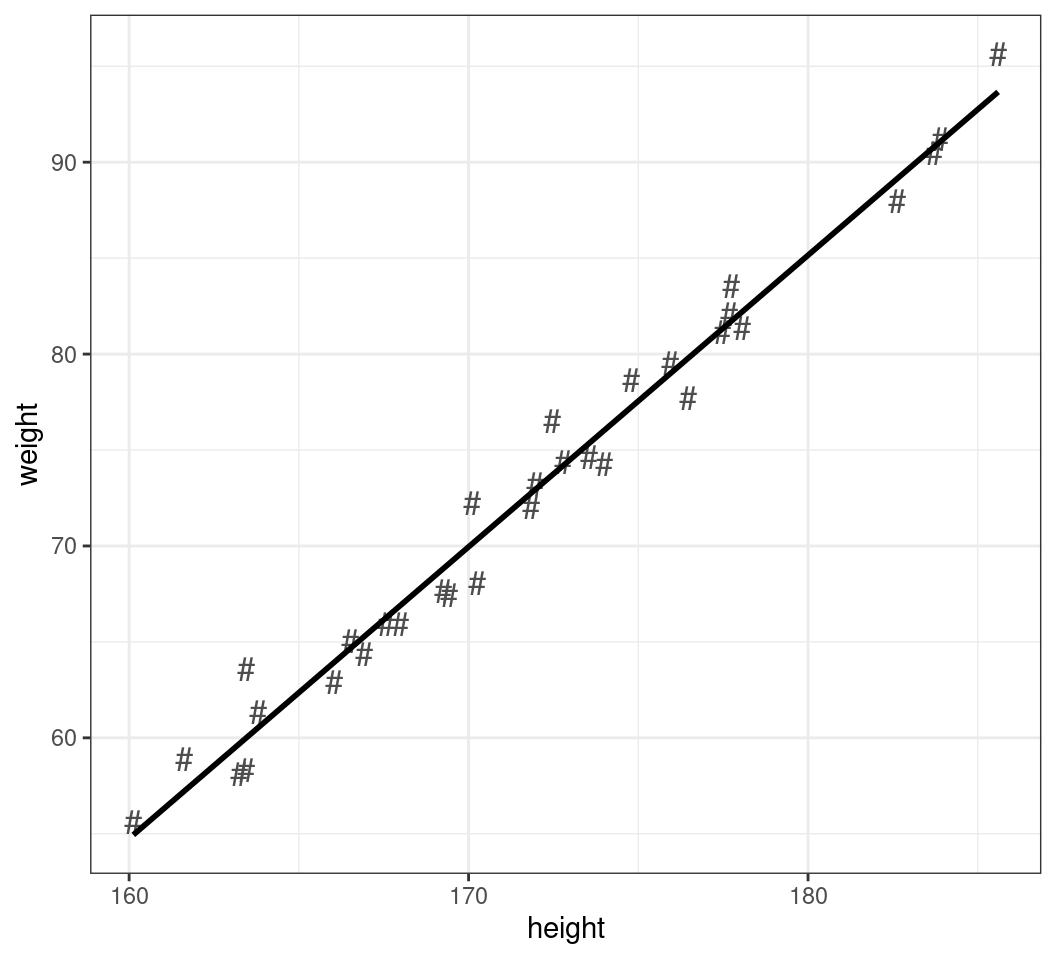
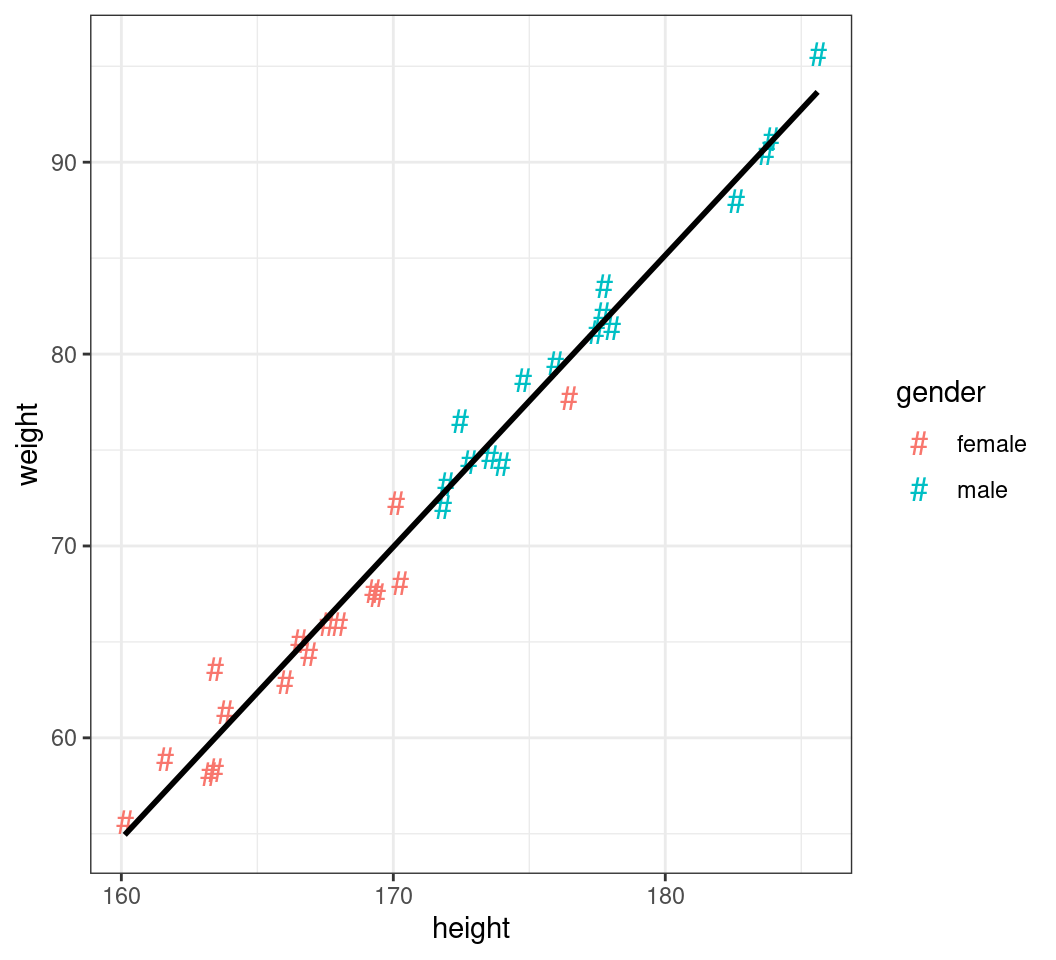
Changing Shapes
- Similarly for the points:
colour = "grey30"- The range of shapes is visible using
?pch - For shapes 21-25 colour is outline, fill is the internal colour
- The range of shapes is visible using
Changing Shapes
- We can also set any character to be the point
shape- Additional parameters include
size,alpha
- Additional parameters include
Parameters Inside or Outside aes()
- Parameters set outside
aes()will over-ride anything insideaes()- We have globally set colour to depend on
gender - This is overridden by both
geom_point()andgeom_smooth()
- We have globally set colour to depend on
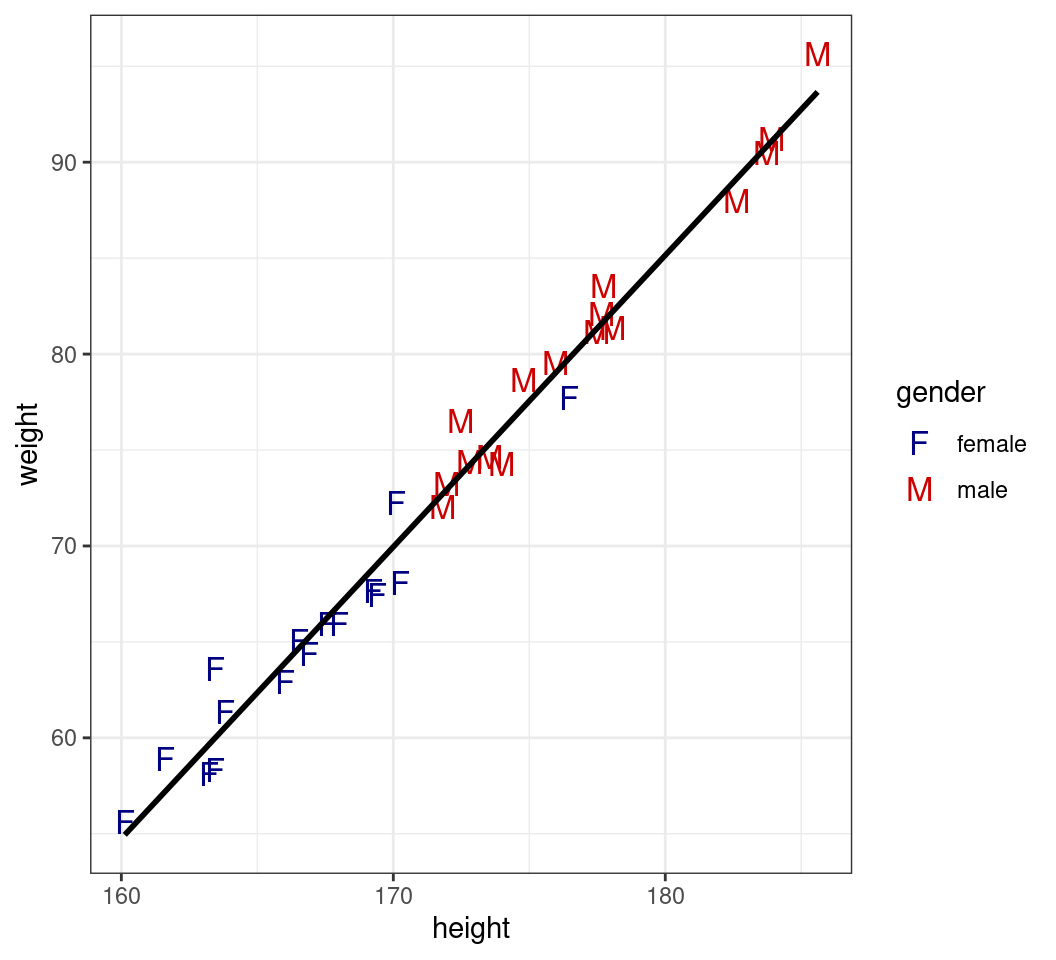
Parameters Inside or Outside aes()
- Now remove the colour from
geom_point()- Inherits from the
aes()withinggplot()
- Inherits from the
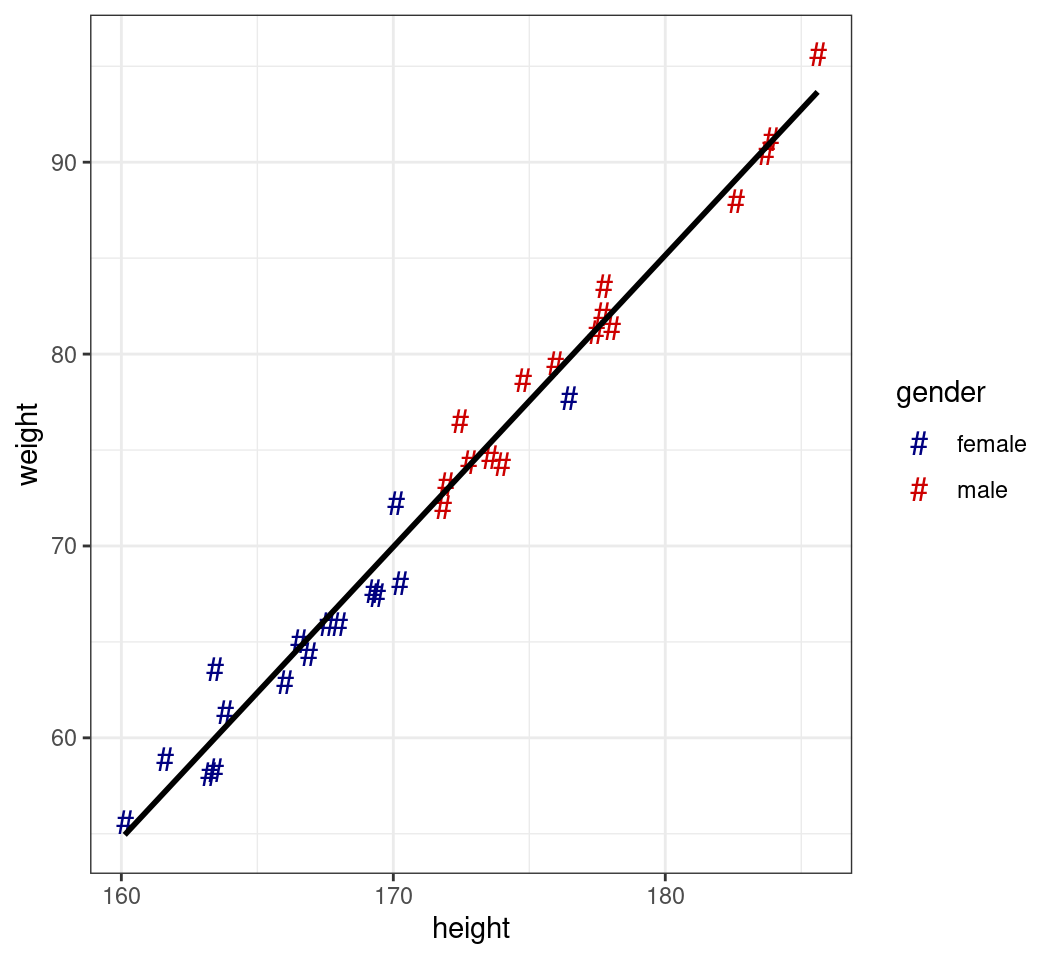
Custom Scales
- Providing specific colours can take a vector
- Can be named for greater control
Custom Scales
- Likewise for shapes
- Values are applied in the same order as the legend
Adding Statistics
- Sometimes we might wish to add summary statistics to plots
- We can create on the fly inside a geom_
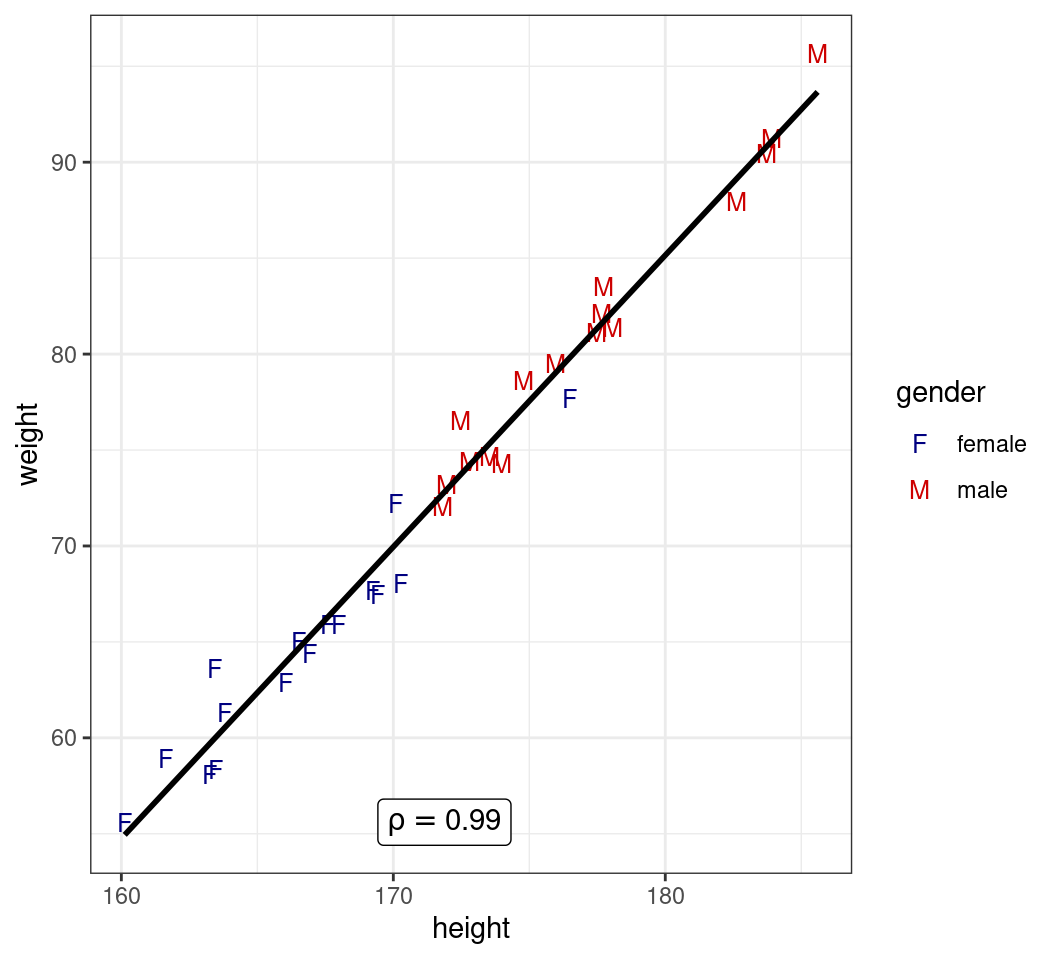
transport |>
ggplot(aes(height, weight)) +
geom_point(
aes(colour = gender, shape = gender),
size = 3
) +
geom_smooth(
method = "lm", se = FALSE,
colour = "black"
) +
geom_label(
aes(label = label),
data = . %>% ## Only the magrittr works here
summarise(
cor = cor(weight, height),
height = mean(height),
## Specify a position manually
# height = 165,
weight = min(weight),
) %>%
mutate(
label = paste("rho ==", round(cor, 2))
),
parse = TRUE
) +
scale_colour_manual(
values = c(
female = "navyblue", male = "red3"
)
) +
scale_shape_manual(
values = c("F", "M")
)
Using Additional Packages
- There are multiple options for adding other labels
Using Additional Packages
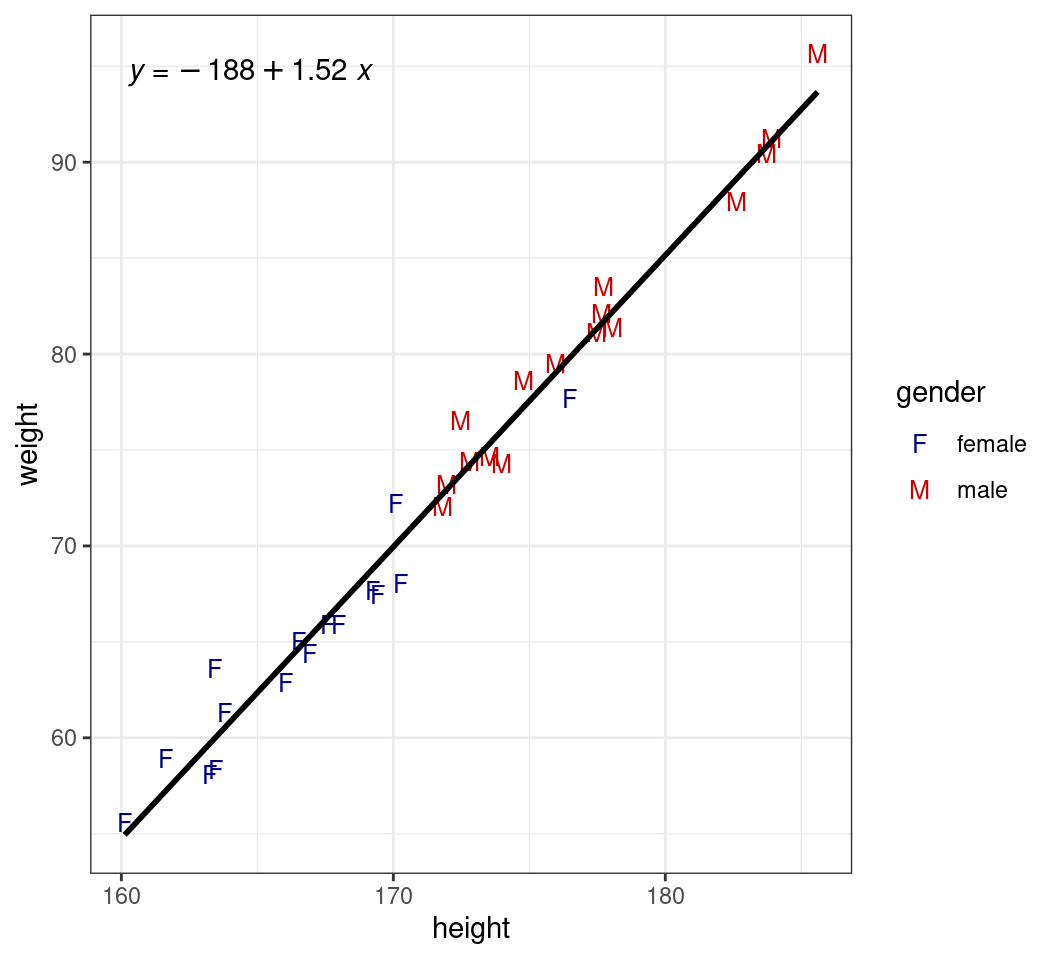
stat_poly_eq()can add \(R^2\), adjusted \(R^2\) or regression equations
transport |>
ggplot(aes(height, weight)) +
geom_point(
aes(colour = gender, shape = gender),
size = 3
) +
geom_smooth(
method = "lm", se = FALSE,
colour = "black"
) +
stat_poly_eq(use_label("eq")) +
scale_colour_manual(
values = c(
female = "navyblue", male = "red3"
)
) +
scale_shape_manual(
values = c("F", "M")
)
Multiple Regression Equations
- Combining with facets can provide multiple equations
transport |>
ggplot(aes(height, weight)) +
geom_point(
aes(colour = gender, shape = gender),
size = 3
) +
geom_smooth(
method = "lm", se = FALSE,
colour = "black"
) +
stat_poly_eq(use_label("eq")) +
facet_wrap(~method) +
scale_colour_manual(
values = c(
female = "navyblue", male = "red3"
)
) +
scale_shape_manual(
values = c("F", "M")
)
Modifying Data
Prior To Plotting
Summary Plots
- So far we’ve just plotted the complete dataset
- We did use
%>%inside ageom_*to find a correlation…
- We did use
- We can also use our
tidyversetools to create summaries to plot- E.g. Barplots of the mean with error bars
Summary Plots
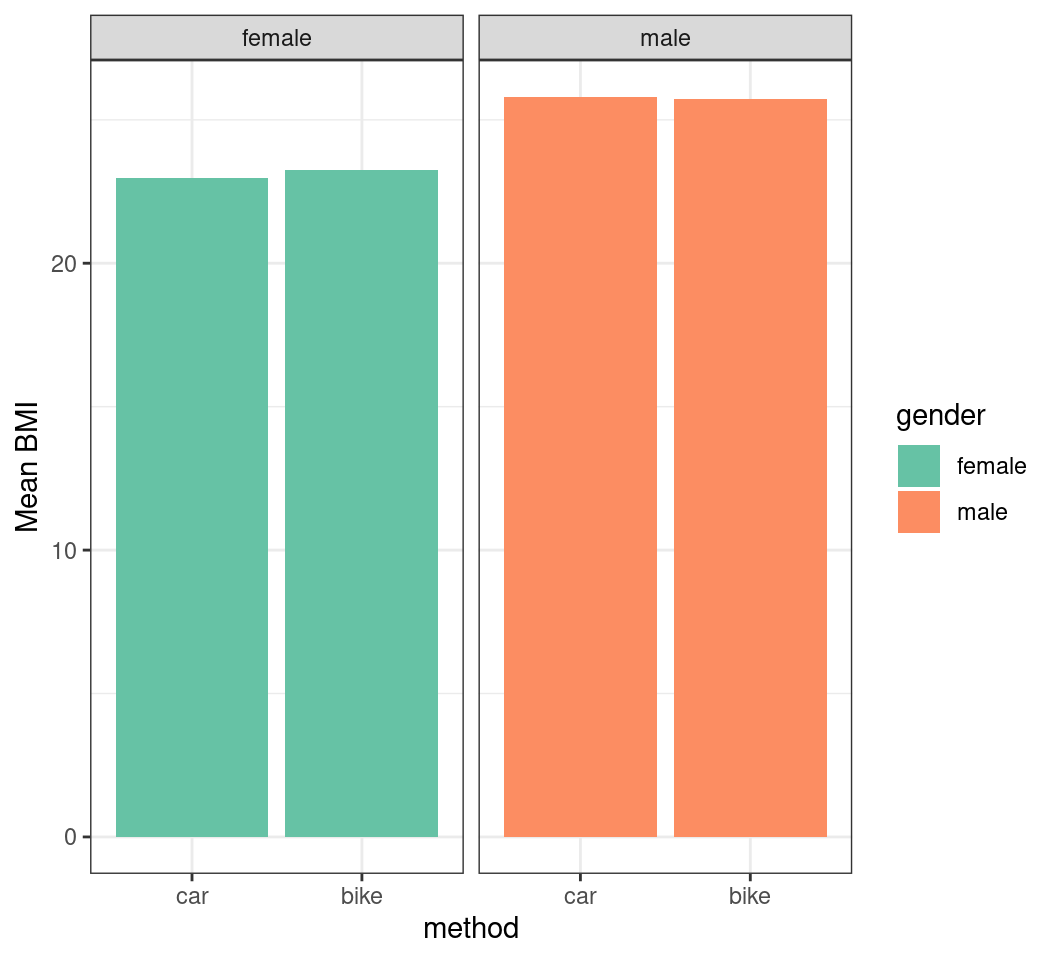
- To create a barplot of the mean BMI across all groups
- Need the mean BMI for each group
- Also the standard deviation
Creating a Barplot
- Now we can create a barplot using
geom_col()
Adding Error Bars
- We add error bars using
geom_errorbar()
transport |>
summarise(
mn_bmi = mean(BMI), sd_bmi = sd(BMI),
.by = c(method, gender)
) |>
ggplot(aes(method, mn_bmi, fill = gender)) +
geom_col() +
geom_errorbar(
aes(
ymin = mn_bmi - sd_bmi,
ymax = mn_bmi + sd_bmi
),
width = 0.2
) +
facet_wrap(~gender) +
labs(y = "Mean BMI") +
scale_fill_brewer(palette = "Set2") 
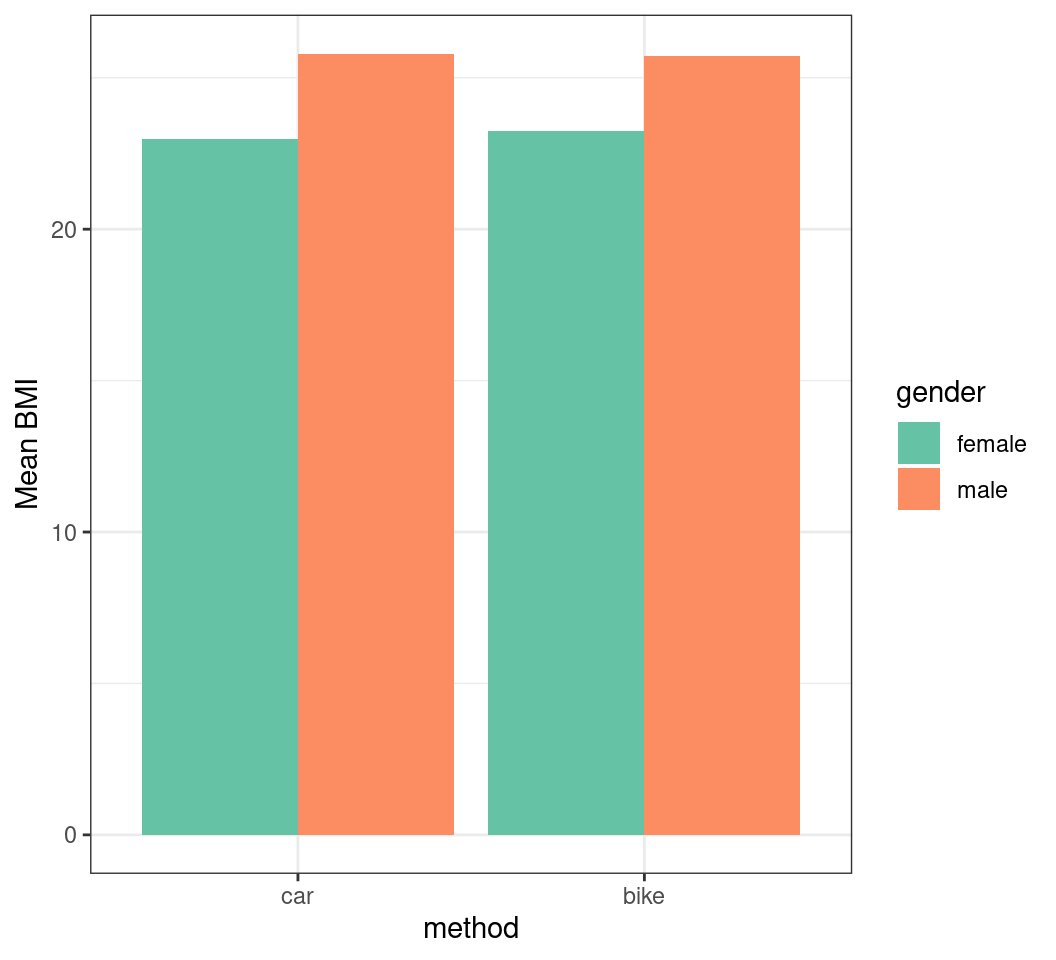
Adding Error Bars
- If we choose not to facet it’s much trickier
- Let’s hide the error bars to see why
Adding Error Bars
- Adding
position = "dodge"separates the bars horizontally - What is the x co-ordinate for each bar now?
- What is the width of each bar?
Plotting with Factors
- Let’s try coerce factors to integers
# A tibble: 4 × 6
method gender mn_bmi sd_bmi method_int gender_int
<fct> <fct> <dbl> <dbl> <int> <int>
1 car female 23.0 1.14 1 1
2 bike female 23.3 0.961 2 1
3 bike male 25.7 1.47 2 2
4 car male 25.8 0.799 1 2Plotting with Factors
- Each factor level is assigned an integer starting at 1
- These were how ggplot originally placed the x co-ordinates
- Can we figure out our x-co-ordinates yet?
- I guesstimated that each bar had a width of 0.45
- The centre of each bar is
method_int\(\pm\) 0.225
- The centre of each bar is
- I also wish it was easier
- Maybe I just haven’t found the right package/function yet
Plotting with Factors
# A tibble: 4 × 7
method gender mn_bmi sd_bmi method_int gender_int x
<fct> <fct> <dbl> <dbl> <int> <int> <dbl>
1 car female 23.0 1.14 1 1 0.775
2 bike female 23.3 0.961 2 1 1.78
3 bike male 25.7 1.47 2 2 2.22
4 car male 25.8 0.799 1 2 1.23 Plotting with Factors
transport |>
summarise(
mn_bmi = mean(BMI), sd_bmi = sd(BMI),
.by = c(method, gender)
) |>
mutate(
method_int = as.integer(method),
x = case_when(
gender == "female" ~ method_int - 0.225,
gender == "male" ~ method_int + 0.225,
)
) |>
ggplot(aes(method, mn_bmi, fill = gender)) +
geom_col(position = "dodge") +
geom_errorbar(
aes(
x = x,
ymin = mn_bmi - sd_bmi,
ymax = mn_bmi + sd_bmi
),
width = 0.1
) +
labs(y = "Mean BMI") +
scale_fill_brewer(palette = "Set2") 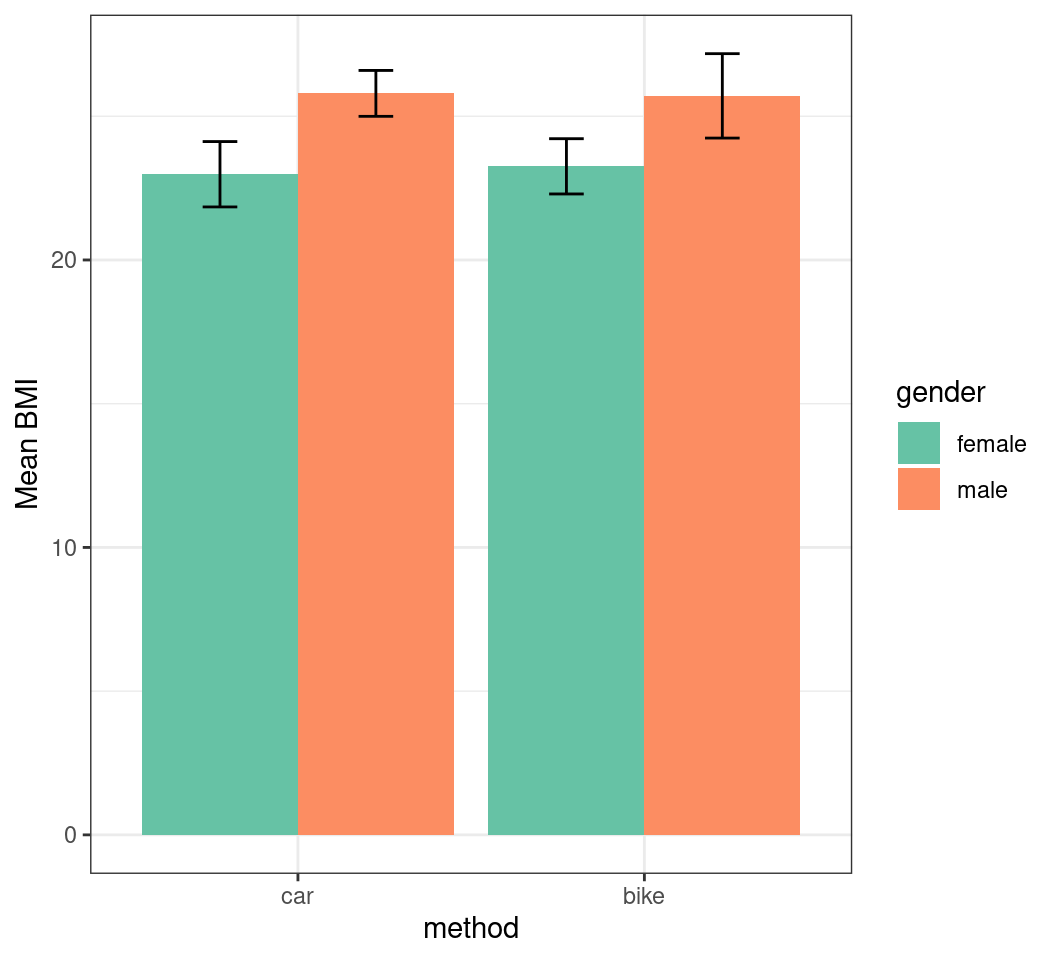
Modifying Axes
- That space at the bottom of the y-axis bother me
- By default axes expand the data by ~5% of the range
- We can control the expansion
- Using multiplicative scaling (continuous data)
- Using Additive Scaling (discrete data)
- The relevant function is called
expansion()- Is passed to the
expandargument insidescale_x/y_*()
- Is passed to the
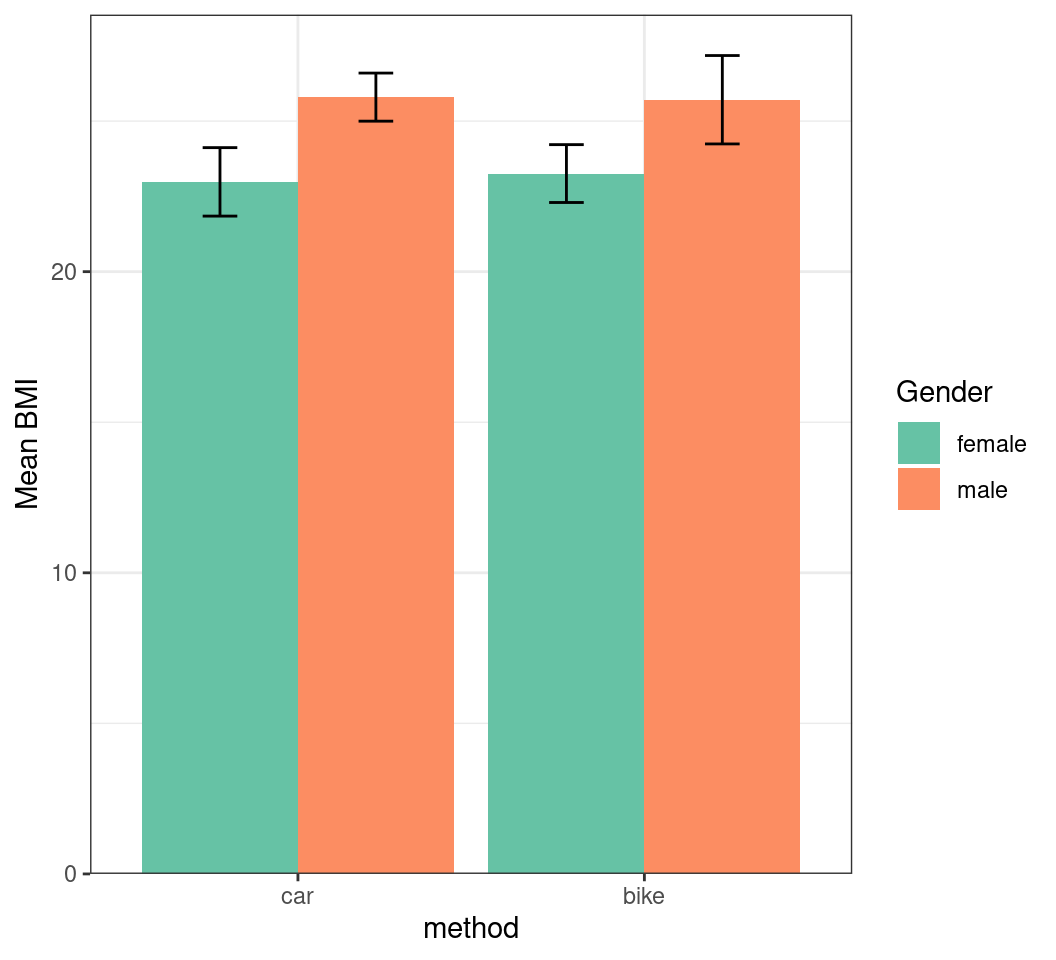
Modifying Axes
- We also label them using the
nameargument
transport |>
summarise(
mn_bmi = mean(BMI), sd_bmi = sd(BMI),
.by = c(method, gender)
) |>
mutate(
method_int = as.integer(method),
x = case_when(
gender == "female" ~ method_int - 0.225,
gender == "male" ~ method_int + 0.225,
)
) |>
ggplot(aes(method, mn_bmi, fill = gender)) +
geom_col(position = "dodge") +
geom_errorbar(
aes(
x = x,
ymin = mn_bmi - sd_bmi,
ymax = mn_bmi + sd_bmi
), width = 0.1
) +
scale_fill_brewer(
palette = "Set2", name = "Gender"
) +
scale_y_continuous(
expand = expansion(c(0, 0.05)),
name = "Mean BMI"
)
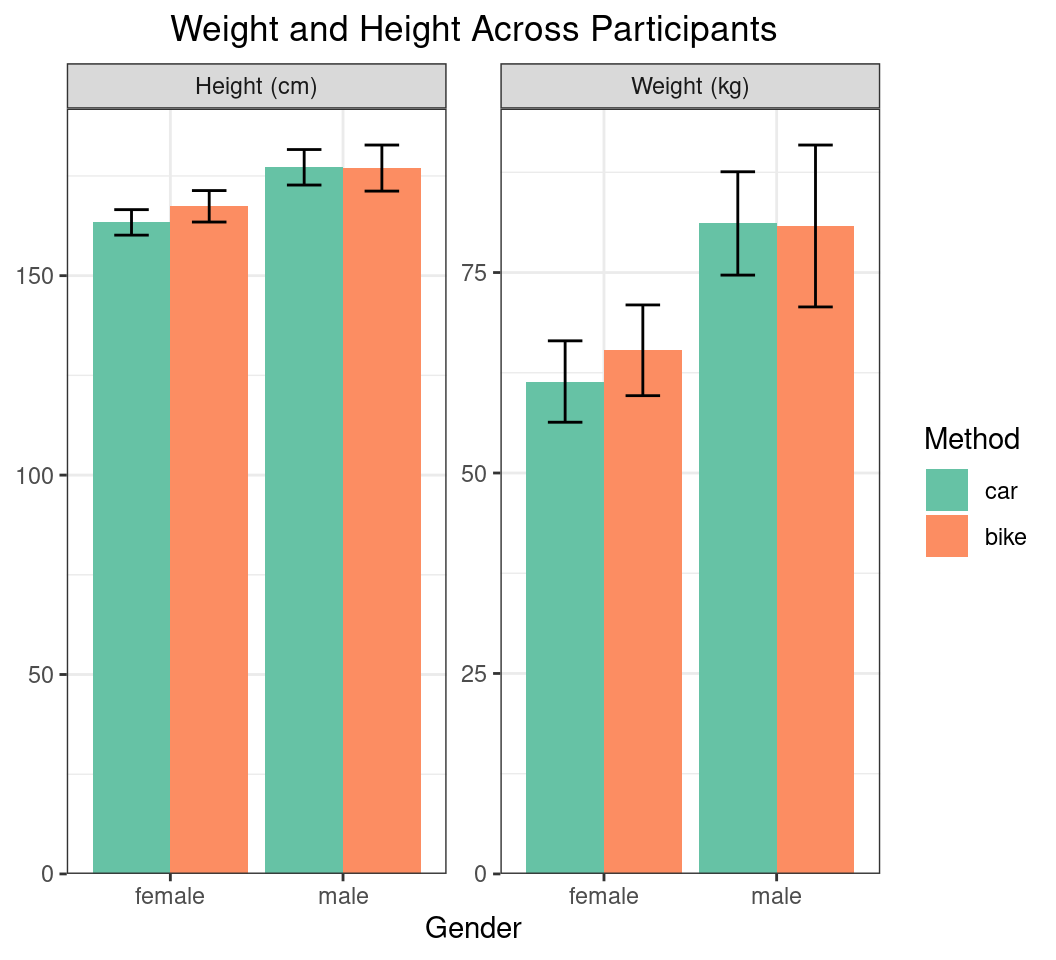
More Complex Summaries
- So far we only visualised the mean BMI values across the dataset
- Let’s try and repeat for both weight and height on the same figure
- May not be brilliant in reality
transport |>
summarise(
across(
ends_with("ght"),
.fns = c(mn = mean, sd = sd)
),
.by = c(method, gender)
) # A tibble: 4 × 6
method gender weight_mn weight_sd height_mn height_sd
<fct> <fct> <dbl> <dbl> <dbl> <dbl>
1 car female 61.4 5.08 163. 3.20
2 bike female 65.3 5.66 167. 3.95
3 bike male 80.8 10.1 177. 5.77
4 car male 81.1 6.44 177. 4.45
Can we get the means into one column and sd into another?
Reshaping Data
pivot_longer()enables shifting from wide to long form- Choose the columns to run down the page
- Original column names go into one column
- All values go into another
# A tibble: 16 × 4
method gender name value
<fct> <fct> <chr> <dbl>
1 car female weight_mn 61.4
2 car female weight_sd 5.08
3 car female height_mn 163.
4 car female height_sd 3.20
5 bike female weight_mn 65.3
6 bike female weight_sd 5.66
7 bike female height_mn 167.
8 bike female height_sd 3.95
9 bike male weight_mn 80.8
10 bike male weight_sd 10.1
11 bike male height_mn 177.
12 bike male height_sd 5.77
13 car male weight_mn 81.1
14 car male weight_sd 6.44
15 car male height_mn 177.
16 car male height_sd 4.45Reshaping Data
- We can now
separate()the values innameinto = c("stat", "type")defines the new column names
# A tibble: 16 × 5
method gender stat type value
<fct> <fct> <chr> <chr> <dbl>
1 car female weight mn 61.4
2 car female weight sd 5.08
3 car female height mn 163.
4 car female height sd 3.20
5 bike female weight mn 65.3
6 bike female weight sd 5.66
7 bike female height mn 167.
8 bike female height sd 3.95
9 bike male weight mn 80.8
10 bike male weight sd 10.1
11 bike male height mn 177.
12 bike male height sd 5.77
13 car male weight mn 81.1
14 car male weight sd 6.44
15 car male height mn 177.
16 car male height sd 4.45Reshaping Data
- The opposite of
pivot_longer()ispivot_wider()
# A tibble: 8 × 5
method gender stat mn sd
<fct> <fct> <chr> <dbl> <dbl>
1 car female weight 61.4 5.08
2 car female height 163. 3.20
3 bike female weight 65.3 5.66
4 bike female height 167. 3.95
5 bike male weight 80.8 10.1
6 bike male height 177. 5.77
7 car male weight 81.1 6.44
8 car male height 177. 4.45
Now we have something we can use for barplots with errorbars
Reshaping Data
- For the sake of brevity going forward, save that as
summ_df
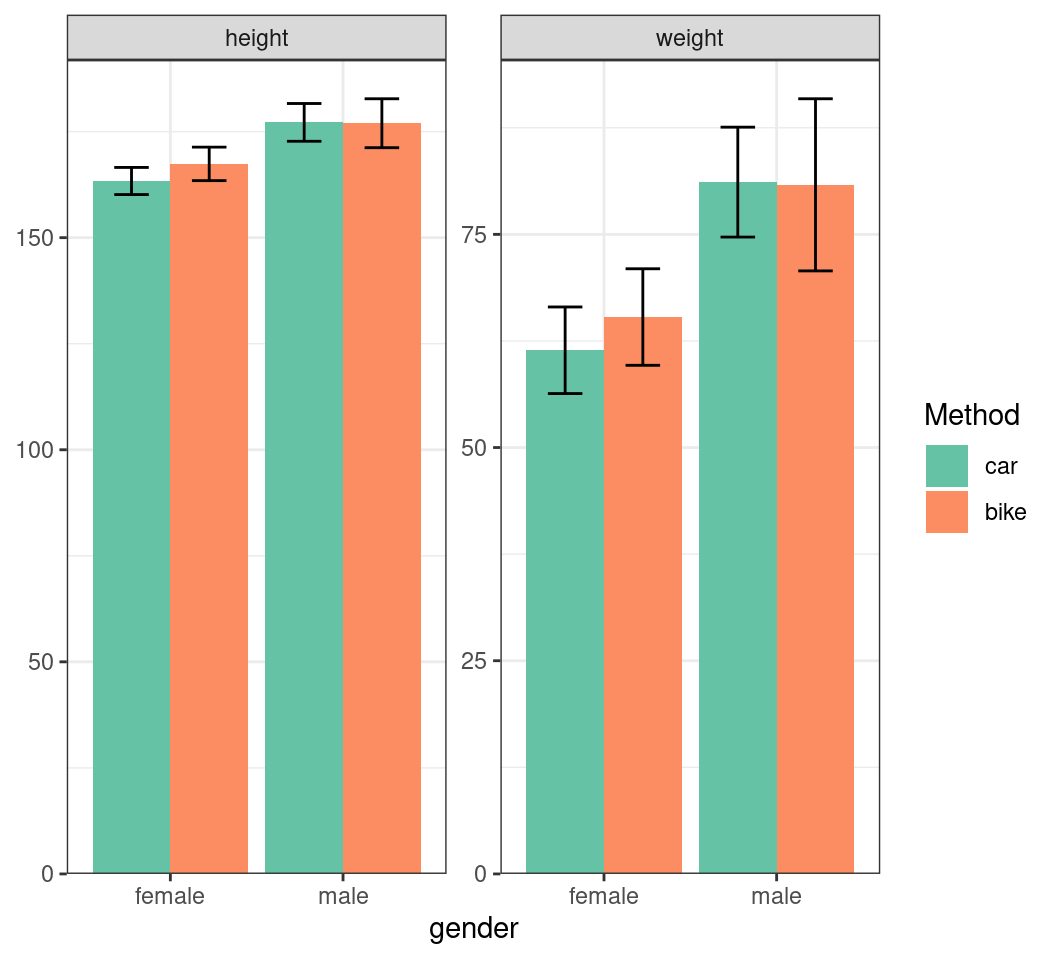
Plotting Multiple Summaries
- Hide the y-axis name using
name = c()
summ_df |>
mutate(
gender_int = as.numeric(gender),
x_bar = case_when(
method == "car" ~ gender_int - 0.225,
method == "bike" ~ gender_int + 0.225
)
) |>
ggplot(aes(gender, mn, fill = method)) +
geom_col(position = "dodge") +
geom_errorbar(
aes(
x = x_bar,
ymin = mn - sd, ymax = mn + sd
),
width = 0.2
) +
facet_wrap(~stat, scales = "free_y") +
scale_fill_brewer(
palette = "Set2", name = "Method"
) +
scale_y_continuous(
expand = expansion(c(0, 0.05)),
name = c()
) 
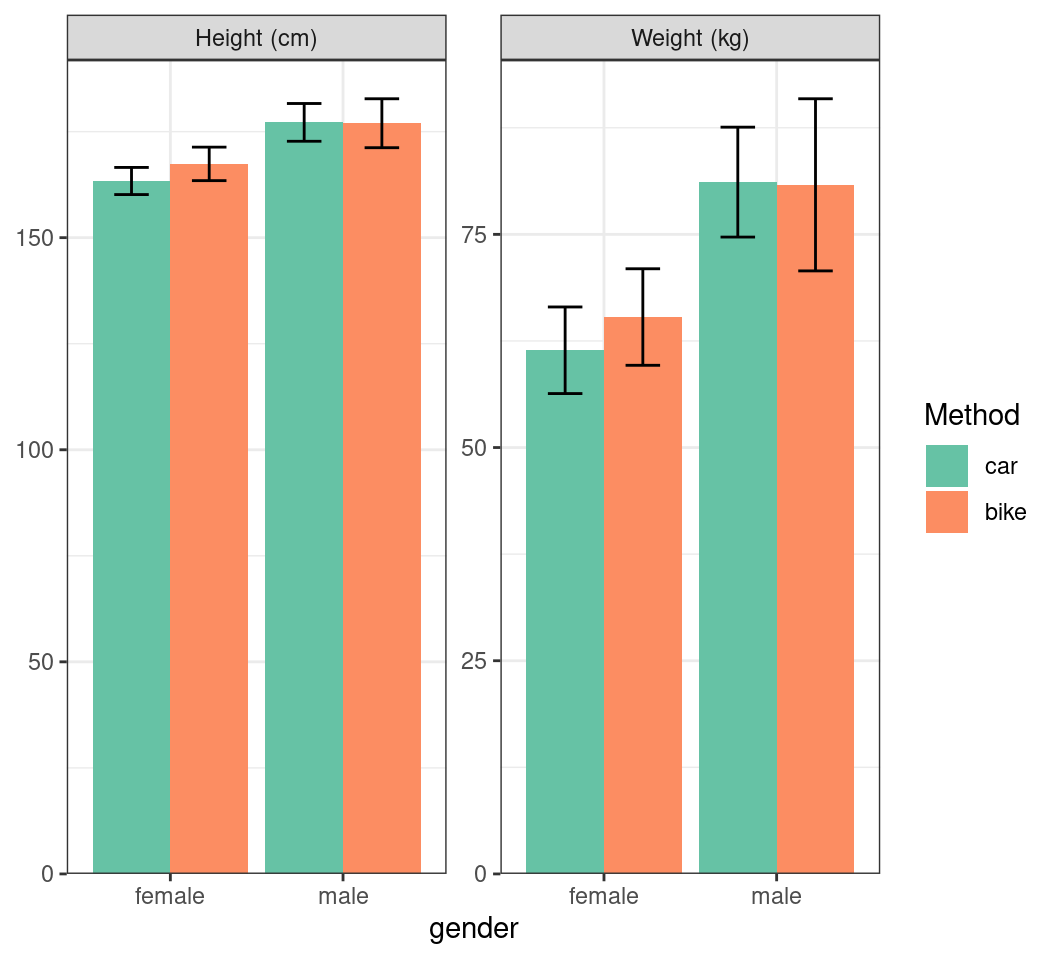
Plotting Multiple Summaries
- Add the units to the facet names
summ_df |>
mutate(
gender_int = as.numeric(gender),
x_bar = case_when(
method == "car" ~ gender_int - 0.225,
method == "bike" ~ gender_int + 0.225
),
stat = stat |>
str_replace("height", "Height (cm)") |>
str_replace("weight", "Weight (kg)")
) |>
ggplot(aes(gender, mn, fill = method)) +
geom_col(position = "dodge") +
geom_errorbar(
aes(
x = x_bar,
ymin = mn - sd, ymax = mn + sd
),
width = 0.2
) +
facet_wrap(~stat, scales = "free_y") +
scale_fill_brewer(
palette = "Set2", name = "Method"
) +
scale_y_continuous(
expand = expansion(c(0, 0.05)), name = c()
) 
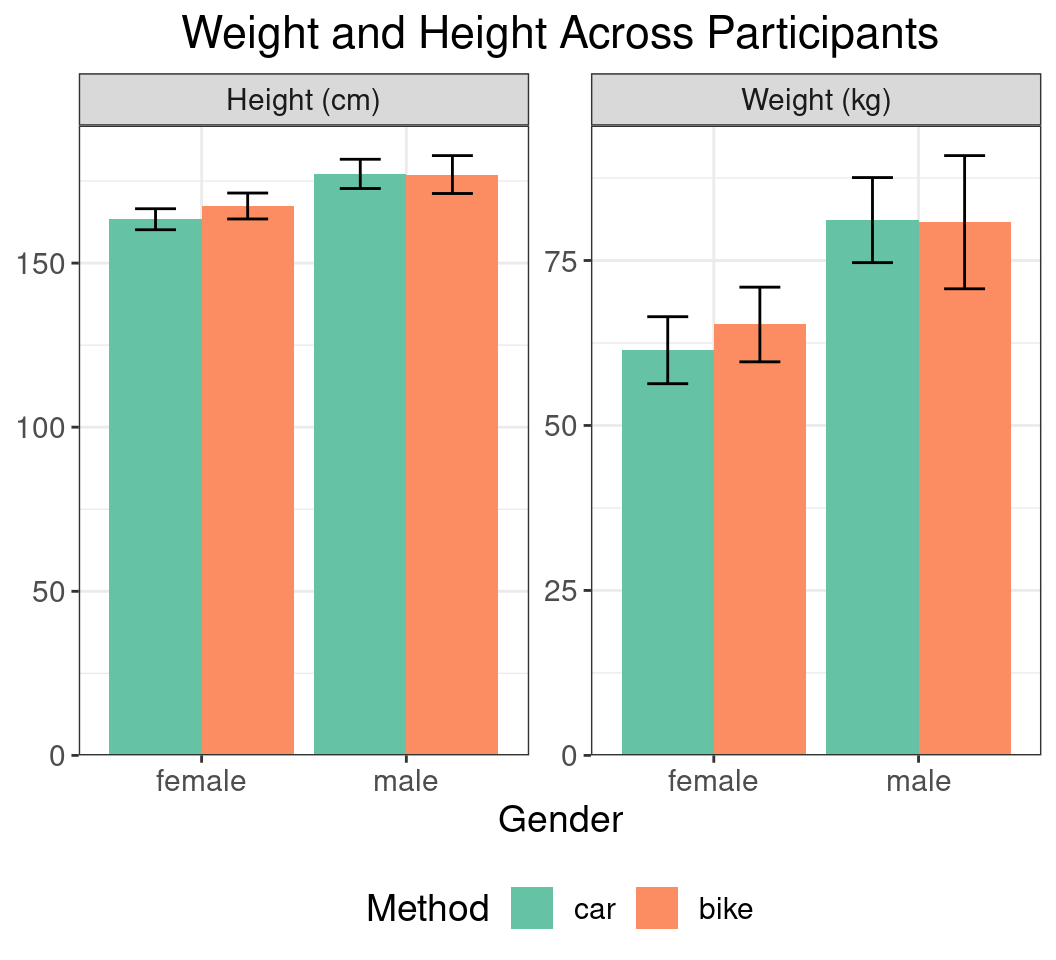
Themes
Using Themes
- The overall plot appearance can be set using
theme() - A bit overwhelming, but very customisable
- Let’s save our existing plot a
p- Can be drawn again at anytime by entering
p - Can still be modified on on-the-fly
- Can be drawn again at anytime by entering
p <- summ_df |>
mutate(
gender_int = as.numeric(gender),
x_bar = case_when(
method == "car" ~ gender_int - 0.225,
method == "bike" ~ gender_int + 0.225
),
stat = stat |>
str_replace("height", "Height (cm)") |>
str_replace("weight", "Weight (kg)")
) |>
ggplot(aes(gender, mn, fill = method)) +
geom_col(position = "dodge") +
geom_errorbar(
aes(
x = x_bar,
ymin = mn - sd, ymax = mn + sd
),
width = 0.2
) +
facet_wrap(~stat, scales = "free_y") +
xlab("Gender") +
scale_fill_brewer(
palette = "Set2", name = "Method"
) +
scale_y_continuous(
expand = expansion(c(0, 0.05)), name = c()
) Using Themes
- If we try to add a title, it’s aligned left

Using Themes
- Most
theme()elements have their own modification functions- text:
element_text() - lines:
element_line() - rectangles:
element_rect()
- text:
- All can be removed using
element_blank
- The plot title needs
element_text()
Using Themes
Using Themes
Using Themes
Using Themes
- Hide the background grid & rotates x-axis text
Exporting Plots
- Making plots in
Ris nice
\(\implies\) How do we get them into our paper??!!!
ggsave()will save the last plot- The file extension will determine the format
- Can be
png,jpg,pdf,svg,tiffetc width&heightdefault to inches but can be changed
- Getting font sizes right can be infuriating
- Always add the save after you create the plot
- Open immediately and check the font sizes
Closing Comments
- Can now (hopefully) make the figures for our next paper
ggplot2is very powerful \(\implies\) takes a long time to master- Getting data structured correctly is an important part
- Note that once we loaded data \(\implies\) never modified
- We saved four objects
cols,transport,summ_df,p- The last two were only to fit the code on slides
- This keeps a clean workspace
- No need for
transport,transport1,transport1_modetc - Very beneficial for reproducibility
- No need for