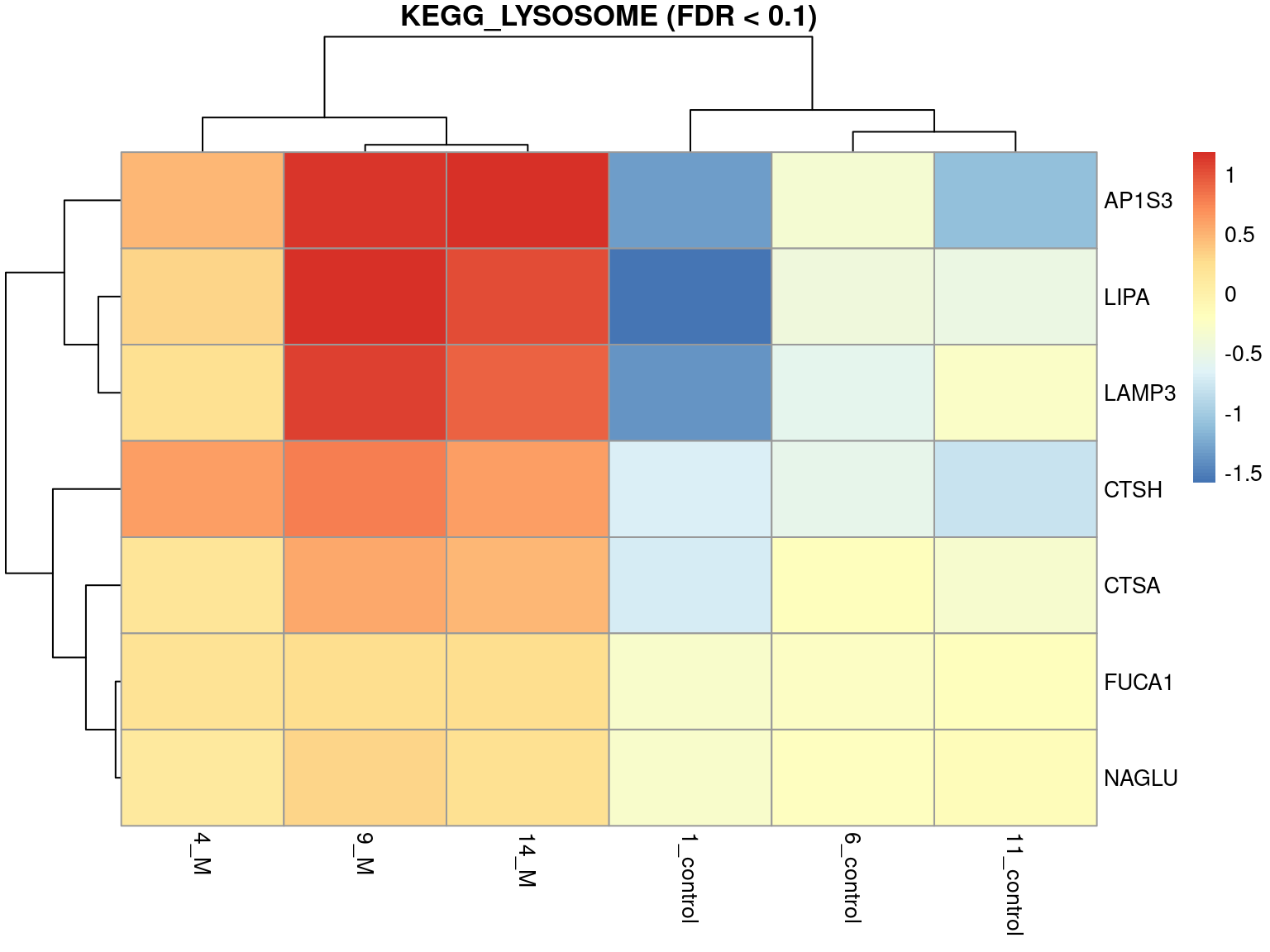
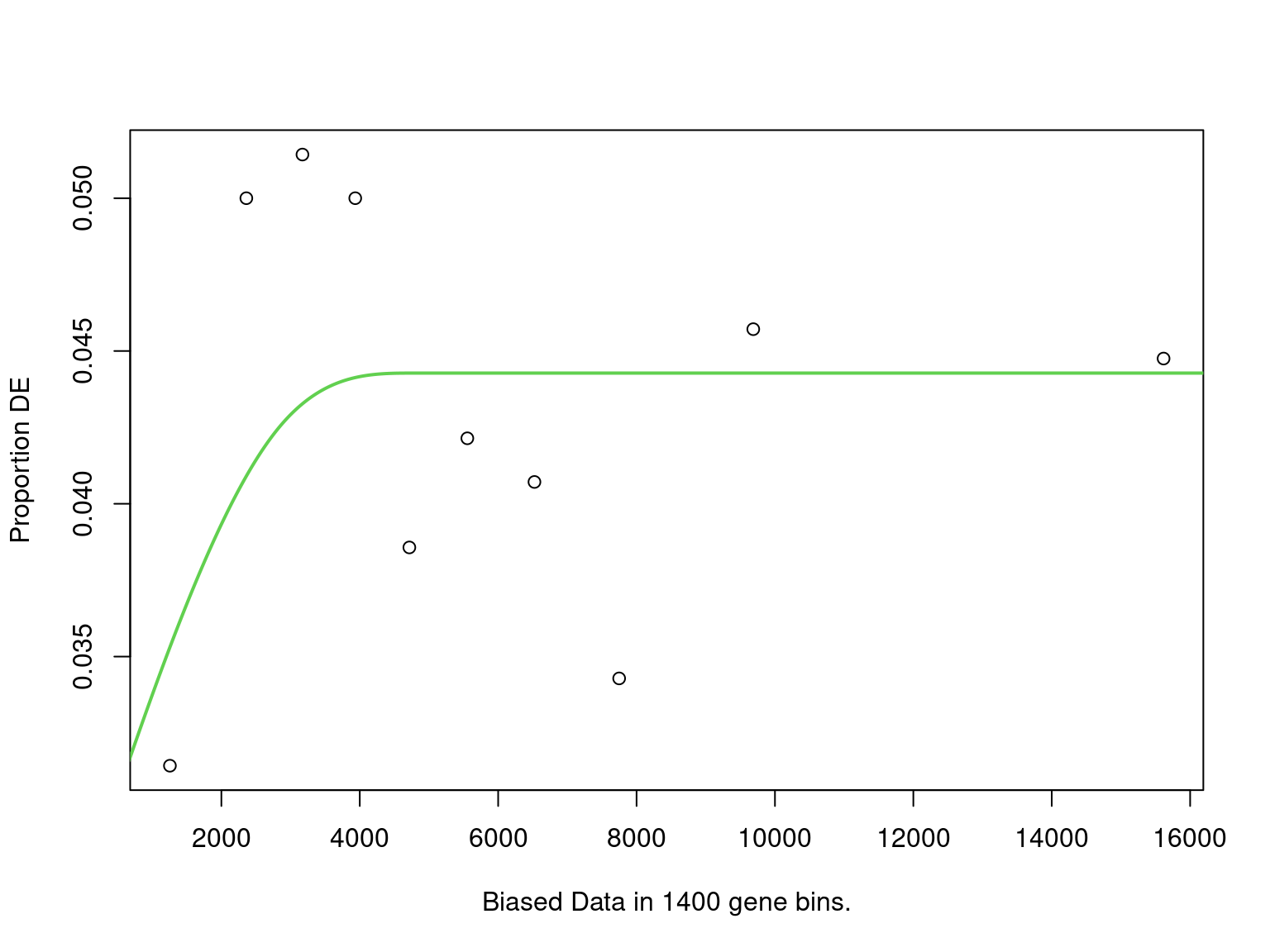
Rows: 16,283
Columns: 15
$ gs_cat <chr> "C2", "C2", "C2", "C2", "C2", "C2", "C2", "C2", "C2…
$ gs_subcat <chr> "CP:KEGG", "CP:KEGG", "CP:KEGG", "CP:KEGG", "CP:KEG…
$ gs_name <chr> "KEGG_ABC_TRANSPORTERS", "KEGG_ABC_TRANSPORTERS", "…
$ gene_symbol <chr> "ABCA1", "ABCA10", "ABCA12", "ABCA13", "ABCA2", "AB…
$ entrez_gene <int> 19, 10349, 26154, 154664, 20, 21, 24, 23461, 23460,…
$ ensembl_gene <chr> "ENSG00000165029", "ENSG00000154263", "ENSG00000144…
$ human_gene_symbol <chr> "ABCA1", "ABCA10", "ABCA12", "ABCA13", "ABCA2", "AB…
$ human_entrez_gene <int> 19, 10349, 26154, 154664, 20, 21, 24, 23461, 23460,…
$ human_ensembl_gene <chr> "ENSG00000165029", "ENSG00000154263", "ENSG00000144…
$ gs_id <chr> "M11911", "M11911", "M11911", "M11911", "M11911", "…
$ gs_pmid <chr> "", "", "", "", "", "", "", "", "", "", "", "", "",…
$ gs_geoid <chr> "", "", "", "", "", "", "", "", "", "", "", "", "",…
$ gs_exact_source <chr> "hsa02010", "hsa02010", "hsa02010", "hsa02010", "hs…
$ gs_url <chr> "http://www.genome.jp/kegg/pathway/hsa/hsa02010.htm…
$ gs_description <chr> "ABC transporters", "ABC transporters", "ABC transp…