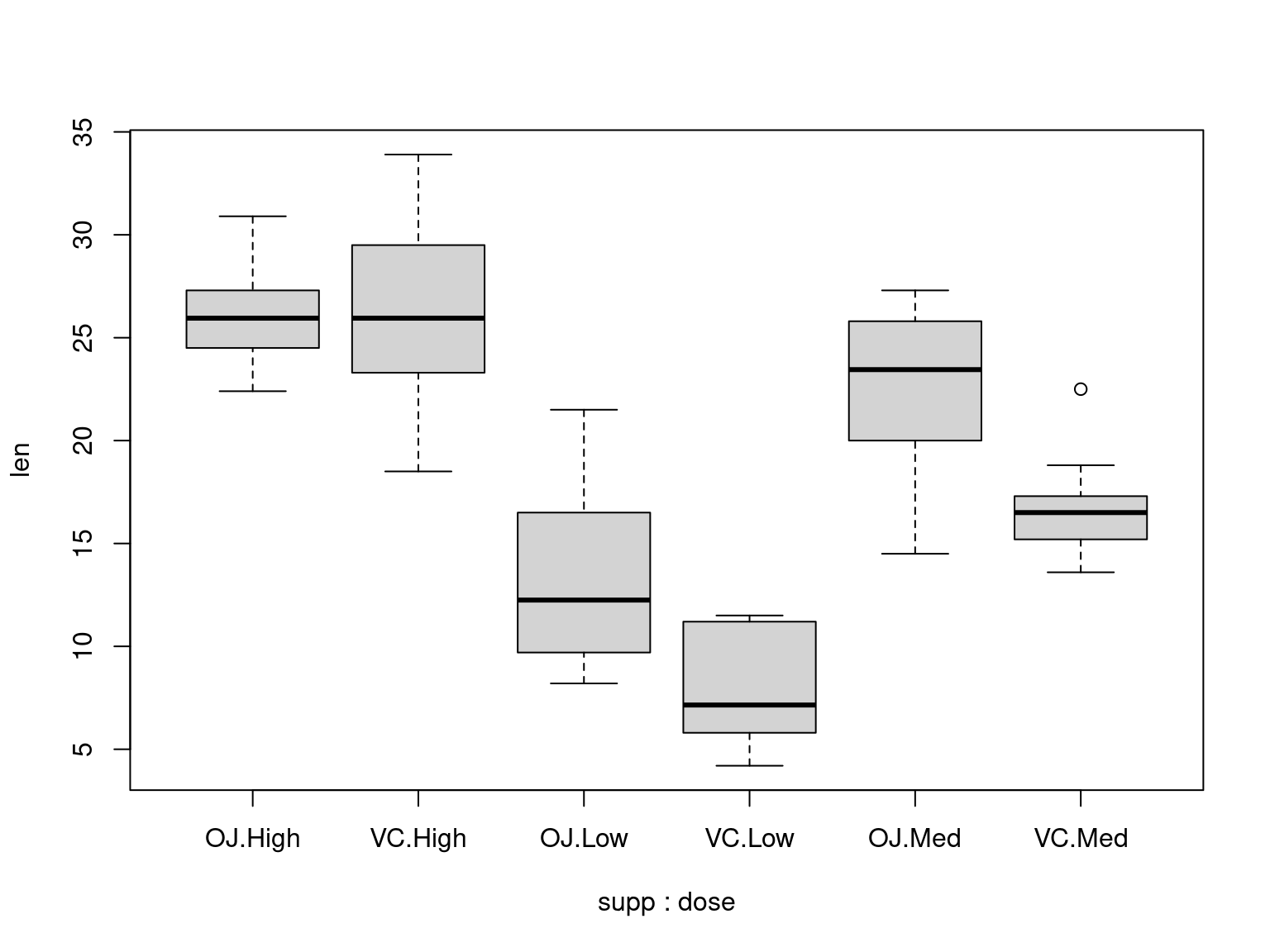
## Look at all of the odontoblast lengths
pigs$len [1] 4.2 11.5 7.3 5.8 6.4 10.0 11.2 11.2 5.2 7.0 16.5 16.5 15.2 17.3 22.5
[16] 17.3 13.6 14.5 18.8 15.5 23.6 18.5 33.9 25.5 26.4 32.5 26.7 21.5 23.3 29.5
[31] 15.2 21.5 17.6 9.7 14.5 10.0 8.2 9.4 16.5 9.7 19.7 23.3 23.6 26.4 20.0
[46] 25.2 25.8 21.2 14.5 27.3 25.5 26.4 22.4 24.5 24.8 30.9 26.4 27.3 29.4 23.0