cars %>%
ggplot(aes(x = speed, y = dist))Introductory Visualisation
RAdelaide 2024
Dr Stevie Pederson
Black Ochre Data Labs
Telethon Kids Institute
Telethon Kids Institute
July 9, 2024
Visualisation With
ggplot2
The Grammar of Graphics
ggplot2has become the industry standard for visualisation (Wickham 2016)- Core & essential part of the
tidyverse - Developed by Hadley Wickham as his PhD thesis
- An implementation of The Grammar of Graphics (Wilkinson 2005)
- Breaks visualisation into layers
The Grammar of Graphics

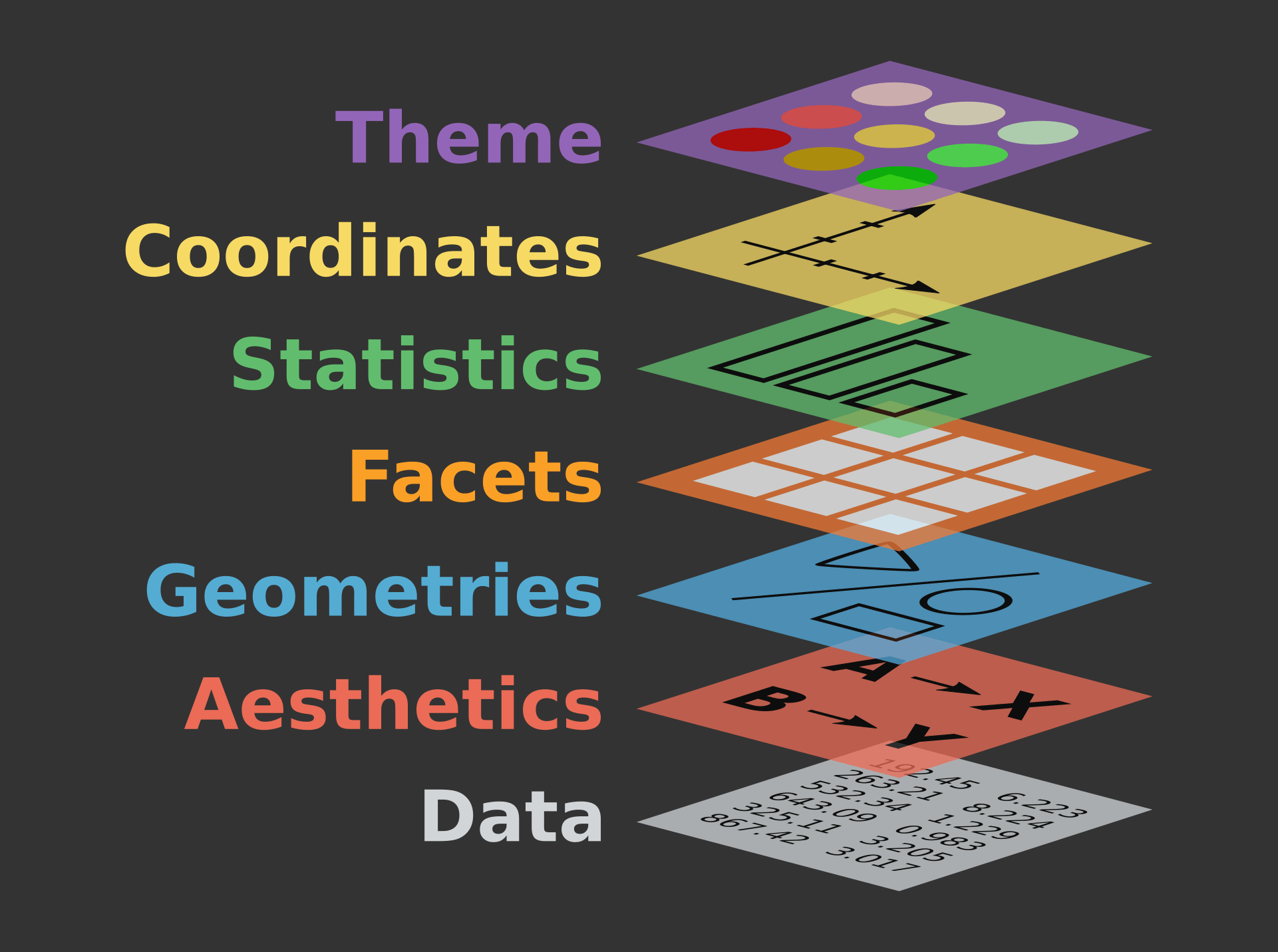
Taken from https://r.qcbs.ca/workshop03/book-en/grammar-of-graphics-gg-basics.html
The Grammar of Graphics
Everything is added in layers
- Data
- Usually a data.frame (or
tibble) - Can be piped in \(\implies\) modify on the fly
- Usually a data.frame (or
- Aesthetics
x&yco-ordinatescolour,fill,shape,size,linetype- grouping & transparency (
alpha)
- Geometric Objects
- points, lines, boxplot, histogram, bars etc
- Facets: Panels within plots
- Statistics: Computed summaries
- Coordinates
- polar, map, cartesian etc
- defaults to cartesian
- Themes: overall layout
- default themes automatically applied
An Initial Example
- Using the example dataset
cars - Two columns:
speed(mph)distanceeach car takes to stop
- We can make a classic
xvsyplot using points
- The predictor (x) would be
speed - The response (y) would be
distance
An Initial Example
- We may as well start by piping our data in
- We have defined the plotting aesthetics
x&y- Don’t need to name if passing in order
- Axis limits match the data
- No geometry has been specified \(\implies\) nothing was drawn
An Initial Example
- To add points, we add
geom_point()after callingggplot()- Adding
+afterggplot()says “But wait! There’s more…”
- Adding
An Initial Example
- To add points, we add
geom_point()after callingggplot()- Adding
+afterggplot()says “But wait! There’s more…”
- Adding
- By default:
- Layer 4: No facets
- Layer 5: No summary statistics
- Layer 6: Cartesian co-ordinate system
- Layer 7: Crappy theme with grey background 🤮
Visualising Our Guinea Pig Data
What visualisations could we produce to inspect pigs?
Creating Our Boxplot
- A starting point might be to choose
doseas the predictor lenwill always be the response variable
Creating Our Boxplot
- To incorporate the supp methods \(\implies\) add a fill aesthetic
colouris generally applied to shape outlines
ggplot2will always separate multiple values/category
Creating Our Boxplot
- We could also separate by supp using
facet_wrap()- Can also set the number of rows/columns
- Only one value/category so no shifting
Layering Geometries
- We’re not restricted to one geometry
- The following will add points after drawing the boxplots
Layering Geometries
geom_jitter()will add a small amount of noise to separate points
Modifying Data Prior to Plotting
doseis a clearly a categorical variable with an order- In
Rthese are known asfactors- Categories referred to as
levels - Will learn in detail in the next session
- Categories referred to as
ggplot()will automatically place character columns in alphanumeric order- Manually set the order by explicitly setting as a
factorwithlevels
Modifying Data Prior to Plotting
- Notice the column is now described as
fct
Modifying Data Prior to Plotting
- Now boxplots will appear in order
Modifying Data Prior to Plotting
- We can also plot quantiles with a few prior steps
- First rank the
lenvalues \(\implies\) turn into quantiles
# A tibble: 60 × 5
len supp dose rank q
<dbl> <chr> <chr> <dbl> <dbl>
1 4.2 VC Low 1 0.0167
2 11.5 VC Low 15 0.25
3 7.3 VC Low 6 0.1
4 5.8 VC Low 3 0.05
5 6.4 VC Low 4 0.0667
6 10 VC Low 11.5 0.192
7 11.2 VC Low 13.5 0.225
8 11.2 VC Low 13.5 0.225
9 5.2 VC Low 2 0.0333
10 7 VC Low 5 0.0833
# ℹ 50 more rowsModifying Data Prior to Plotting
Modifying Data Prior to Plotting
- Now we could colour points by
supp
Different Layers

Modifying Data Prior to Plotting
geom_smooth()will add a line of best fit- Aliases
stat_smooth()
- Aliases
- Automatically chosen but can be
lm,loessorgam
Modifying Geoms
- Any
aestheticset in the call toggplot()is passed to every subsequent layer - We can set aesthetics in a layer-specific manner
- Shifting
colour = supptogeom_point()will only colour points - The line of best fit will now be a single line
Modifying Geoms
Modifying Geoms
- Aesthetics can also be set outside of a call to
aes()
Modifying Geoms
- Geoms are just regular functions with multiple arguments
- The below turns off the
sebands and switches tolm
Choosing Point Shapes
- Shapes have numeric codes in
R - Examples are on the
?pchpage - The default is 19
- Can also be set as an
aesthetic sizecan also work either way
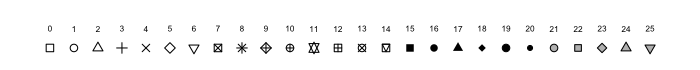
Choosing Point Shapes
Setting Scales
- Default scales are set for x & y axes
scale_x_continuous()&scale_y_continuous()- Only needed when tweaking axis names, limits, labels, breaks etc
- Also set scales for colours, shapes, fill etc
Setting Scales
scale_colour_brewer()allows pre-defined palettes- From the package
RColorBrewer
- From the package
RColorBrewer Palettes

Setting Scales
scale_colour_viridis_b/c/d()- colour-blind friendly palettes
- comes in binned (
_b()), continuous (_c()) or discrete (_d()) - excellent for heatmaps or showing differences across large range
Setting Scales
scale_colour_manual()takes a vector of colours- Vectors are formed using
c() - RStudio helpfully shows you the colour!!!
- Vectors are formed using
Themes
Themes
- We can modify the overall appearance of the plot using
theme() - Set panel colours, fonts, legend position etc
- Hide any features we don’t want
Themes
- To help us focus on the
theme()
\(\implies\) save the plot as the objectp
- We can regenerate the plot by typing it’s name
Themes
ggplot2supplies several complete themes- Applies
theme_grey()by default - Try add
theme_bw()afterp- This is my default
- Try a few others
theme_void(),theme_classic(),theme_minimal()
- Some are for specific use cases
Themes
- We can also modify manually
- Theme elements are modified using
element_*()functions- Text elements use
element_text() - Line elements use
element_line() - Box (or rectangle) elements use
element_rect() - Can disable an element entirely using
element_blank()
- Text elements use
Themes
- The panel background is set using
element_rect()coloursets the rectangle outline colourfillsets the rectangle fill
Themes
- We can set global text parameters using
text = element_text()- family, colour, size, face etc
Themes
- Individual text-based parameters can be set similarly
- Will over-ride any global setting
Themes
- Can also set a theme then modify further
- Enormous range of setting can be controlled here
Themes
- Spend a few minutes playing with the following
- Try commenting out lines or changing values
- Aesthetic names can be set manually using
labs()- Won’t over-write anything set in
scale_x/y_continuous()
- Won’t over-write anything set in
p +
ggtitle("Odontoblast Length in Guinea Pigs") +
labs(colour = NULL) +
theme(
rect = element_rect(fill = "#204080"),
text = element_text(colour = "grey80", family = "Palatino", size = 14),
panel.background = element_rect(fill = "steelblue4", colour = "grey80"),
panel.grid = element_line(colour = "grey80", linetype = 2, linewidth = 1/4),
axis.text = element_text(colour = "grey80"),
legend.background = element_rect(fill = "steelblue4", colour = "grey80"),
legend.key = element_rect(colour = NA),
legend.position = "inside",
legend.position.inside = c(1, 0),
legend.justification = c(1, 0),
plot.title = element_text(hjust = 0.5, face = "bold"),
)Themes

Saving Images
- The simple way is click
Exportin thePlotspane
Saving Images
- I think saving using code is preferable
- Modify an analysis or data \(\implies\) saved figures will also update
- This saves time & ensures reproducibility
Conclusion

A fabulous resource: https://r-graphics.org/
References

Wickham, Hadley. 2016. Ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag New York. https://ggplot2.tidyverse.org.
Wilkinson, Leland. 2005. The Grammar of Graphics. Springer New York, NY. https://doi.org/https://doi.org/10.1007/0-387-28695-0.