pigs <- read_csv("data/pigs.csv") %>%
mutate(
dose = factor(dose, levels = c("Low", "Med", "High")),
supp = factor(supp, levels = c("VC", "OJ"))
)R Markdown
RAdelaide 2024
Telethon Kids Institute
July 10, 2024
R Markdown

Writing Reports Using rmarkdown
rmarkdownis a cohesive way to- Load & wrangle data
- Analyse data, including figures & tables
- Publish everything in a complete report/analysis
- The package
knitris the engine behind this- Replaced the
Sweavepackage about 8-10 years ago
- Replaced the
Extends the markdown language to incorporate R code
Writing Reports Using rmarkdown
- Everything is one document
- Our analysis code embedded alongside our explanatory text
- The entire analysis is performed in a fresh R Environment
- Avoids issues with saving/re-saving Workspaces
- Effectively enforces code that runs sequentially
Starting With Markdown
A Brief Primer on Markdown
- Markdown is a simple and elegant way to create formatted HTML
- Text is entered as plain text
- Formatting usually doesn’t appear on screen (but can)
- The parsing to HTML often occurs using
pandoc
- Often used for Project README files etc.
- Not R-specific but is heavily-used across data-science
- Go to the File drop-down menu in RStudio
- New File -> Markdown File
- Save As
README.md
Editing Markdown
- Section Headers are denoted by on or more
#symbols#is the highest level,##is next highest etc.
- Italic text is set by enclosing text between a single asterisk (
*) or underscore (_)
- Bold text is set by using two asterisks (
**) or two underscores (__)
Editing Markdown
- Dot-point Lists are started by prefixing each line with
-- Next level indents are formed by adding 2 or 4 spaces before the next
-
- Next level indents are formed by adding 2 or 4 spaces before the next
- Numeric Lists are formed by starting a line with
1.- Subsequent lines don’t need to be numbered in order
Editing Markdown
Let’s quickly edit our file so there’s something informative
Enter this on the top line
# RAdelaide 2024
Two lines down add this
## Day 1
Leave another blank line then add
1. Introduction to R and R Studio
2. Importing Data
3. Data Exploration
4. Data Visualisation
Editing Markdown
Underneath the list enter:
**All material** can be found at [the couse homepage](http://blackochrelabs.au/RAdelaide24/)
- Here we’ve set the first two words to appear in bold font
- The section in the square brackets will appear as text with a hyperlink to the site in the round brackets
- Click the
Preview Buttonand an HTML document appears - Note that README.html has also been produced
- Sites like github/gitlab render this automatically
- Obsidian also renders interactively
R Markdown
Writing Reports Using rmarkdown
We can output our analysis directly as:
- HTML
- MS Word Documents
- PDF Documents (If you have \(\LaTeX\) installed)
- Slidy,
ioslidesor PowerPoint presentations
We never need to use MS Word, Excel or PowerPoint again!
Writing Reports Using rmarkdown
- The file suffix is
.Rmd - Include both markdown + embedded
Rcode. - Create all of our figures & tables directly from the data
- Data, experimental and analytic descriptions
- Mathematical/Statistical equations
- Nicely Formatted Results
- Any other information: citations, hyperlinks etc.
Creating an R Markdown document
Let’s create our first rmarkdown document
- Go to the
Filedrop-down menu in RStudio - New File -> R Markdown…
Creating an R Markdown document
- Change the Title to: My First Report
- Change the Author to your preferred name
- Leave everything else as it is & hit OK
- Save the file as
RMarkdownTutorial.Rmd
Looking At The File
A header section is enclosed between the --- lines at the top
- Nothing can be placed before this!
- Uses YAML (YAML Ain’t Markup Language)
- Editing is beyond the scope of this course
- Can set custom
.cssfiles, load LaTeX packages, set parameters etc.
Looking At The File
Lines 8 to 10 are a code chunk
- Chunks always begin with ```{r}
- Chunks always end with ```
- Executed
Rcode goes between these two delineation marks
- Chunk names are optional and directly follow the letter
r- Chunks can also be other languages (
bash,pythonetc.) - Here the
rtells RMarkdown the chunk is anRchunk
- Chunks can also be other languages (
- Other parameters are set in the chunk header (e.g. do we show/hide the code)
- The master reference is here
Looking At The File
Line 12 is a Subsection Heading, starting with ##
- Click the Outline symbol in the top-right of the Script Window
- Chunk names are shown in italics (if set to be shown)
Tools>Global Options>R Markdown- Show in document outline: Sections and All Chunks
- Section Names in plain text
- Chunks are indented within Sections
- By default Sections start with
##- Only the Document Title should be Level 1
#
- Only the Document Title should be Level 1
Getting Help
Check the help for a guide to the syntax.
Help > Markdown Quick Reference
- Increasing numbers of
#gives Section \(\rightarrow\) Subsection \(\rightarrow\) Subsubsection etc. - Bold is set by **Knit** (or __Knit__)
- Italics can be set using a single asterisk/underline: *Italics* or _Italics_
Typewriter fontis set using a single back-tick `Typewriter`
Compiling The Report
The default format is an html_document & we can change this later. Generate the default document by clicking Knit
Compiling The Report
The Viewer Pane will appear with the compiled report (probably)
- Note the hyperlink to the RMarkdown website & the bold typeface for the word Knit
- The R code and the results are printed for
summary(cars) - The plot of
temperatureVs.pressurehas been embedded - The code for the plot was hidden using
echo = FALSE
Compiling The Report
- We could also export this as an MS Word document by clicking the small ‘down’ arrow next to the word Knit.
- By default, this will be Read-Only, but can be helpful for sharing with collaborators.
- Saving as a
.PDFrequires \(\LaTeX\)- Beyond the scope of today
Making Our Own Report
Making Our Own Report
Now we can modify the code to create our own analysis.
- Delete everything in your R Markdown file EXCEPT the header
- We’ll analyse the
pigsdataset - Edit the title to be something suitable
Making Our Own Reports
What do we need for our report?
- Load and describe the data using clear text explanations
- Maybe include the questions being asked by the study
- Create figures which show any patterns, trends or issues
- Perform an analysis
- State conclusions
- Send to collaborators
Making Our Own Reports
- First we’ll need to load the data
- Then we can describe the data
- RMarkdown always compiles from the directory it is in
- File paths should be relative to this
- My “first” real chunk always loads the packages we need
Creating a Code Chunk
Alt+Ctrl+Icreates a new chunk on Windows/LinuxCmd+Option+Ion OSX
- Type
load-packagesnext to the ```{r- This is the chunk name
- Really helpful habit to form
- Enter
library(tidyverse)in the chunk body- We’ll add other packages as we go
Knit…
Dealing With Messages
- The
tidyverseis a little too helpful sometimes- These messages look horrible in a final report
- Are telling us which packages/version
tidyversehas loaded - Also informing us of conflicts (e.g.
dplyr::filterVs.stats::filter) - Can be helpful when running an interactive session
- We can hide these from our report
Dealing With Messages
- Go to the top of your file (below the YAML)
- Create a new chunk
- Name it
setup - Place a comma after
setupand addinclude = FALSE- This will hide the chunk from the report
- In the chunk body add
knitr::opts_chunk$set(message = FALSE)- This sets a global parameter for all chunks
- i.e. Don’t print “helpful” messages
Knit…
Making Our Own Reports
- I like to load all data straight after loading packages
- Gets the entire workflow sorted at the beginning
- Alerts to any problems early
Below the load-packages chunk:
- Create a new chunk
- Name it
load-data - In the chunk body load
pigsusingread_csv()
Describing Data
Now let’s add a section header for our analysis to start the report
- Type
## Data Descriptionafter the header and after leaving a blank line - Use your own words to describe the data
- Consider things like how many participants, different methods, measures we have etc.
60 guinea pigs were given vitamin C, either in their drinking water in via orange juice. 3 dose levels were given representing low, medium and high doses. Odontoblast length was measured in order to assess the impacts on tooth growth
Describing Data
- In my version, I mentioned the study size
- We can take this directly from the data
- Very useful as participants change
nrow(pigs)would give us the number of pigs
Replace the number 60 in your description with `r nrow(pigs)`
Knit…
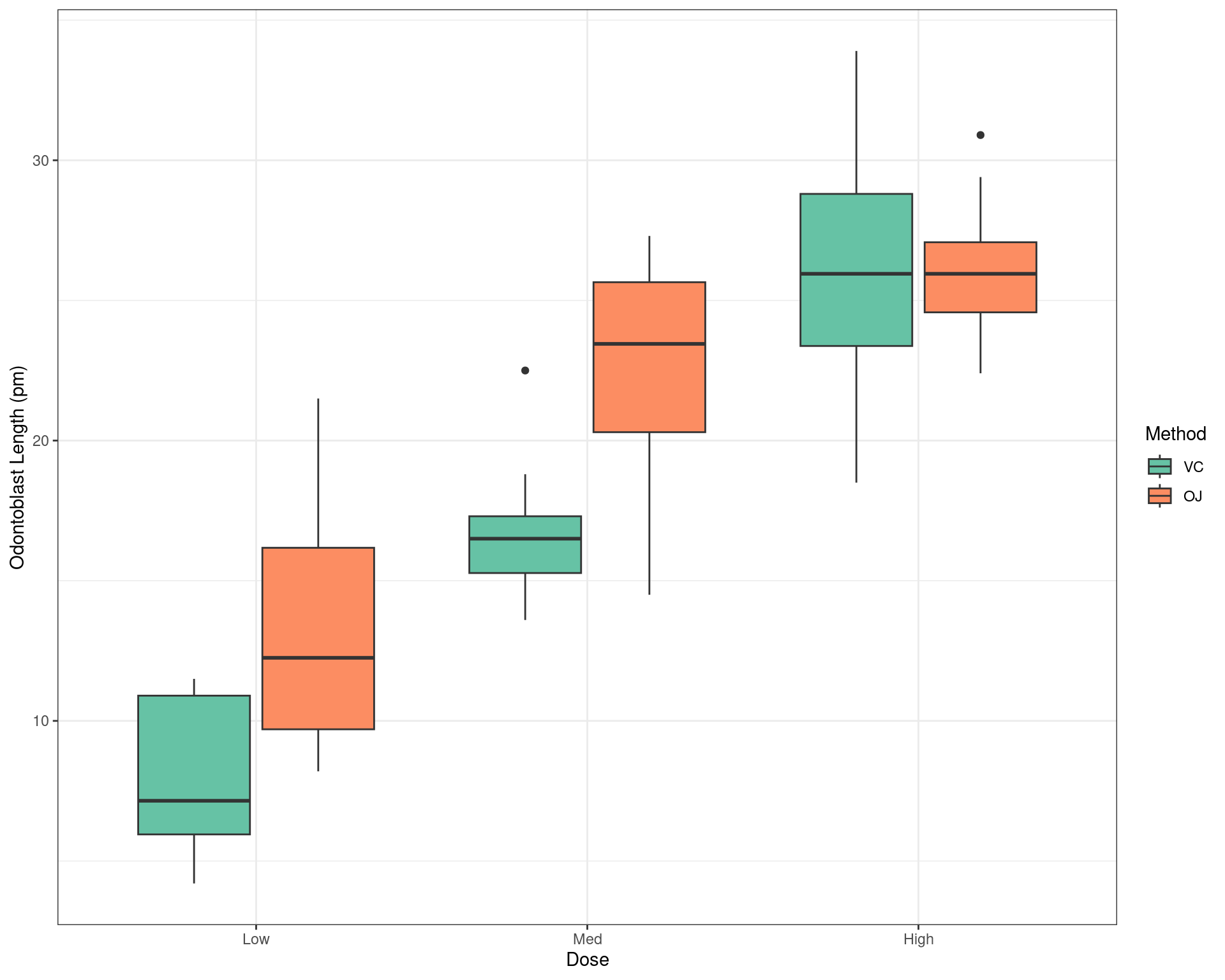
Visualising The Data
- The next step might be to visualise the data using a boxplot
- Start a new chunk with ```{r boxplot-data}
Visualising the Data
- Type a description of the figure in the
fig.capsection of the chunk header- This will need to be placed inside quotation marks
My example text:
Odontoblast length shown by supplement method and dose level
Summarising Data
- Next we might like to summarise the data as a table
- Show group means & standard deviations
- Add the following to a new chunk called
data-summary- I’ve used the HTML code for \(\pm\) (±)
# A tibble: 6 × 4
supp dose n len
<fct> <fct> <int> <chr>
1 VC Low 10 7.98 ±2.75
2 VC Med 10 16.77 ±2.52
3 VC High 10 26.14 ±4.8
4 OJ Low 10 13.23 ±4.46
5 OJ Med 10 22.7 ±3.91
6 OJ High 10 26.06 ±2.66Knit…
Summarising Data
- This has given a tibble output
- We can produce an HTML table using
pander
Producing Tables
pigs %>%
summarise(
n = n(),
mn_len = mean(len),
sd_len = sd(len),
.by = c(supp, dose)
) %>%
mutate(
mn_len = round(mn_len, 2),
sd_len = round(sd_len, 2),
len = paste0(mn_len, " ±", sd_len)
) %>%
dplyr::select(supp, dose, n, len) %>%
rename_with(str_to_title) %>%
pander(
caption = "Odontoblast length for each group shown as mean±SD"
)Producing Tables
| Supp | Dose | N | Len |
|---|---|---|---|
| VC | Low | 10 | 7.98 ±2.75 |
| VC | Med | 10 | 16.77 ±2.52 |
| VC | High | 10 | 26.14 ±4.8 |
| OJ | Low | 10 | 13.23 ±4.46 |
| OJ | Med | 10 | 22.7 ±3.91 |
| OJ | High | 10 | 26.06 ±2.66 |
Analysing Data
- Performing statistical analysis is beyond the scope of today BUT
- The function
lm()is used to perform linear regression
Call:
lm(formula = len ~ supp + dose + supp:dose, data = pigs)
Residuals:
Min 1Q Median 3Q Max
-8.20 -2.72 -0.27 2.65 8.27
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 7.980 1.148 6.949 4.98e-09 ***
suppOJ 5.250 1.624 3.233 0.00209 **
doseMed 8.790 1.624 5.413 1.46e-06 ***
doseHigh 18.160 1.624 11.182 1.13e-15 ***
suppOJ:doseMed 0.680 2.297 0.296 0.76831
suppOJ:doseHigh -5.330 2.297 -2.321 0.02411 *
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 3.631 on 54 degrees of freedom
Multiple R-squared: 0.7937, Adjusted R-squared: 0.7746
F-statistic: 41.56 on 5 and 54 DF, p-value: < 2.2e-16Analysing Data
Analysing Data
| Estimate | Std. Error | t value | Pr(>|t|) | ||
|---|---|---|---|---|---|
| (Intercept) | 7.98 | 1.148 | 6.949 | 4.984e-09 | * * * |
| suppOJ | 5.25 | 1.624 | 3.233 | 0.002092 | * * |
| doseMed | 8.79 | 1.624 | 5.413 | 1.463e-06 | * * * |
| doseHigh | 18.16 | 1.624 | 11.18 | 1.131e-15 | * * * |
| suppOJ:doseMed | 0.68 | 2.297 | 0.2961 | 0.7683 | |
| suppOJ:doseHigh | -5.33 | 2.297 | -2.321 | 0.02411 | * |
| Observations | Residual Std. Error | \(R^2\) | Adjusted \(R^2\) |
|---|---|---|---|
| 60 | 3.631 | 0.7937 | 0.7746 |
Creating Summary Tables
- Multiple other packages exist for table creation
- All do some things brilliantly, none does everything
panderis a good all-rounder- Tables are very simplistic
- Also enables easy in-line results
Creating Summary Tables
- To use other packages, \(\implies\)
broom::tidy()- Will convert
lm()output to atibble - This can be passed to other packages which make HTML / \(\LaTeX\) tables
- Will convert
# A tibble: 6 × 5
term estimate std.error statistic p.value
<chr> <dbl> <dbl> <dbl> <dbl>
1 (Intercept) 7.98 1.15 6.95 4.98e- 9
2 suppOJ 5.25 1.62 3.23 2.09e- 3
3 doseMed 8.79 1.62 5.41 1.46e- 6
4 doseHigh 18.2 1.62 11.2 1.13e-15
5 suppOJ:doseMed 0.680 2.30 0.296 7.68e- 1
6 suppOJ:doseHigh -5.33 2.30 -2.32 2.41e- 2Creating Summary Tables
reactablecreates amazing looking tablesDTalso creates fantastic tables- Less flexible with formatting
- Allows simple downloading to
csv,xlsetc.
gtis popular with somextableis excellent for \(\LaTeX\) output
Complete the Analysis
After you’re happy with the way your analysis looks
- A good habit is to finish with a section called
Session Info - Add a code chunk which calls the R command
sessionInfo()
So far we’ve been compiling everything as HTML, but let’s switch to an MS Word document. We could email this to our collaborators, or upload to Google docs
Summary
This basic process is incredibly useful
- We never need to cut & paste anything between R and other documents
- Every piece of information comes directly from our R analysis
- We can very easily incorporate new data as it arrives
- Source data is never modified
- Creates reproducible research
- Highly compatible with collaborative analysis & version control (Git)